HTML
-
Baculoviruses are a group of insect-specific, enveloped DNA viruses that are widely used as biocontrol agents against insect pests and as vectors for foreign protein expression. Two progeny phenotypes, the budded virus (BV) and the occlusion-derived virus (ODV), are produced during the biphasic replication cycle of the baculovirus. BVs are responsible for cell-cell systemic infection, whereas ODVs embedded in occlusion bodies (OBs) are essential for horizontal transmission, and initiate oral infection in larval midgut epithelial cells (Slack J M et al., 2007). After being ingested by a susceptible host, OBs dissociate under the alkaline conditions (pH 8.7 to 11) within the host midgut, and release ODVs (Ohkawa T et al., 2005). ODVs bind to unknown receptors and enter by direct membrane fusion with the microvilli of midgut epithelium cells. Multiple proteins known as 'per os infectivity factors' (PIFs) have been found to be essential in baculoviruses for successful establishment of oral infection, including P74 (PIF0) (Haas-Stapleton E J et al., 2004; Kuzio J et al., 1989), PIF1 (Ohkawa T et al., 2005), PIF2 (Ohkawa T et al., 2005), PIF3 (Ohkawa T et al., 2005), PIF4 (Fang M G et al., 2009; Huang H C et al., 2012), PIF5 (ODV-E56) (Harrison R L et al., 2010; Sparks W O et al., 2011; Xiang X W et al., 2011) and PIF6 (Nie Y C et al., 2012). The pif genes are conserved in baculoviral genomes and they are also found in nudiviruses, polydnaviruses and other invertebrate viruses (Burke G R et al., 2013). Thus, a detailed functional investigation of PIFs would facilitate an understanding of the entry processes used by baculoviruses and other invertebrate DNA viruses, and whether these might be evolutionarily conserved.
P74 was the first PIF to be reported (Kuzio J et al., 1989). It has been identified as mediating ODV-specific binding with midgut columnar cell microvilli (Haas-Stapleton E J et al., 2004). In Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV), P74 is anchored onto the ODV envelope by two transmembrane (TM) domains at its C terminus, whilst its N terminus is exposed on the ODV outer surface (Slack J M et al., 2001). A model for the two-step cleavage of P74 during its activation has been proposed, according to which P74 is first cleaved by ODV-associated host alkaline protease, and then digested by trypsin from the host midgut (Peng K et al., 2011). In AcMNPV, the first step of digestion appears to be the generation of fragment(s) of 34 kDa, as detected by Western blotting (Peng K et al., 2011). When incubated with host proteins from the midgut, soluble AcMNPV P74 expressed using a baculoviral vector system can be digested, releasing a smaller N-terminal fragment of about 25 kDa (Slack J M and Lawrence S D, 2005), which might be the product of secondary cleavage. Many studies have focused attention upon the involvement of P74 in the oral infectivity of ODV, and upon the interactions of P74 with host proteins (Haas-Stapleton E J et al., 2004; Yao L G et al., 2004; Zhou W K et al., 2005). In the case of AcMNPV, interactions of P74 with host protein(s) may mediate ODV-specific binding with microvilli (Yao L G et al., 2004; Zhou W K et al., 2005). Furthermore, a truncated P74 lacking the C-terminal TM domains retains activity as a PIF (Slack J M et al., 2010). A monoclonal antibody (mAb) against the N-terminus of AcMNPV P74, N25-8C, has been raised and this has facilitated studies of the cleavage of P74, and of its oral infectivity (Faulkner P et al., 1997; Slack J M and Lawrence S D, 2005).
The Helicoverpa armigera nucleopolyhedrovirus (HearNPV) has been used successfully as a viral pesticide against cotton worm (Sun X L and Peng H Y, 2007), and its genome has been sequenced (Chen X W et al., 2001). In the present study, we have generated three mAbs, using a truncated form of HearNPV P74 (P74△3TM) as the primary immunogen, and have evaluated their properties using Western blotting and immunofluorescence microscopy analyses. The results have revealed the linear recognition regions of 20D9 and 21E1, and have demonstrated that 20D9 and 20F9 can be used for detecting native P74 expressed in HearNPV-infected cells. The results will facilitate future studies on the mechanisms of proteolysis of P74 and on its role in the per os infectivity of HearNPV P74.
-
Cells of Helicoverpa zea cell line Hz-AM1 were maintained in Grace medium (Invitrogen, USA), supplemented with 10 % fetal bovine serum. The HearNPV vHaBac-egfp-ph (Song J J et al., 2008), was used and was propagated in Hz-AM1 cells. The vector pMAL-C2x (New England Biolabs, USA) and the E.coli strain TB1 were used for the expression of recombinant proteins, with the exception of truncated P74 (residues 2-219), which was expressed using pET-32a and E.coli strain BL21.
-
Based on previous investigations of the proteolytic cleavage of AcMNPV P74 (Peng K et al., 2011; Slack J M et al., 2008), a truncated HearNPV P74 (2-460 aa) lacking all three TM domains, P74△3TM, was constructed and further dissected at the proposed cleavage sites into three non-overlapping segments, designated N, M and C (Fig. 1). Segment N was then dissected into peptide fragments Ⅰ, Ⅱ, Ⅲ and Ⅳ, such that there was an overlap of 25 amino acids between each pair of adjacent peptides (Fig. 1). Fragments Ⅲ and Ⅳ were further divided into three and four fragments, respectively, such that between each pair of adjacent peptides, there was an overlap of eight amino acids. Each truncated peptide was fused with fragment Ⅰ at its C terminus. The resulting chimeric P74 derivatives were designated Ⅲ F1-F3 and Ⅳ F1-F4 (Fig. 1). Finally, Ⅲ F3 was dissected into Ⅲ F3 (1)-(5) and Ⅳ F4 was dissected into Ⅳ F4 (1)-(7), which in each case were fused with fragment Ⅰ at the C terminus (Fig. 1).
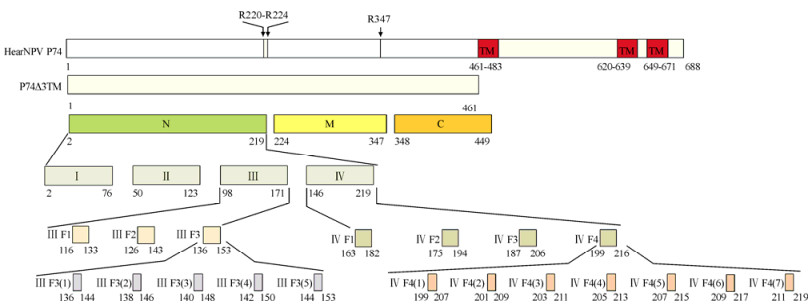
Figure 1. Schematic diagram of HearNPV P74 fragments. The full-length structure of HearNPV P74 is shown at the top. The two putative cleavage regions of P74 are indicated by arrowheads. The three conserved C-terminal transmembrane domains predicted by the TMHMM 2.0 server (http://www.cbs.dtu.dk/services/TMHMM/) are shown in red and indicated as TM. The names of the truncated and chimeric P74 fragments are indicated within, or to the left of, the corresponding rectangles. The location of each fragment within HearNPV P74 is indicated beneath.
The gene fragments encoding the above peptides were PCR-amplified using the primer pairs listed in Table 1. All PCR-amplified DNAs, except for that encoding N, were cloned into pMAL-C2x between BamH Ⅰ and Hind Ⅲ sites and expressed in E.coli strain TB1. DNA encoding the N fragment was cloned instead into pET-32a and expressed in E.coli strain BL21. Proteins were expressed and purified according to a standard protocol (NEB).
Fragementsa Primersb, c, d P74△3TM F CGCGAATTCTTCATGTCGAATATCATTTATATAC R GCGAAGCTTTTATTTAATTGCTATTTTAGTCATC N F GCCGGATTCTTCATGTCGAATACATTTATATACA R CGGCTCGGGCAATAGGCTTCGTTGAAAAAAA M F CGCGGATCCCTAGGCGATACTGTACTTGTC R CCCAAGCTTGCGCGTTGCCATCTATCCGTTG C F CGCGGATCCCGTTCCTTACAACCGAACGG R CCCAAGCTTGCCGCCGTAACGGTTTTAATAGC Ⅰ F CGCGGATCCTTCATGTCGAATATCATTTATATAC R CCCAAGCTTGCGTAAAAATCATCATTGT TAGCCG Ⅱ F CGCGGATCCTTCATACCGAAATGGCGGG R CCCAAGCTTGCCGATGTTTGTGTATACCCGAA Ⅲ F CGCGGATCCGAAAGCATGAGTTGTTATCCG R CCCAAGCTTGCTGTCAAAGTGTCCACTATAATAC Ⅳ F CGCGGATCCGGTGCCGAAAATGAAATACAAA R CCCAAGCTTGCGCAATAGGCTTCGTTGAAAAAA Ⅲ F1 F CGCGGATCCCAGTTCGGGTATACACAAACATCGGAAACGGCAGTGGCGTACGCTCAACCCGCATTCATGTCGAATATCATTTATATAC Ⅲ F2 F CGCGGATCCGCAGTGGCGTACGCTCAACCCGCATGCTACAATTTGGACAGGGCGGCGGCGGTGTTCATGTCGAATATCATTTATATAC Ⅲ F3 F CGCGGATCCAATTTGGACAGGGCGGCGGCGGTGCGCGACGGTGCCGAAAATGAAATACAAACGTTCATGTCGAATATCATTTATATAC Ⅳ F1 F CGCGGATCCAAATGTATTATAGTGGACACTTTGACAAAAATGTATTTGAACACTCCCTATTTGCGTACCCAACCTACTGTACCTACGCC Ⅳ F2 F CGCGGATCCTTGAACACTCCCTATTTGCGTACCGATGACCATTTGATACAGGGCGTTGATGATGTGCCCCAACCTACTGTACCTACGCC Ⅳ F3 F CGCGGATCCATACAGGGCGTTGATGATGTGCCCGGATTCAATGTGACAAACGATACGGATCAACTTTTTCAACCTACTGTACCTACGCC Ⅳ F4 F CGCGGATCCACAAACGATACGGATCAACTTTTTCCCGAAAGATTCGAAGGTTTTTTCAACGAAGCCTATTGCCAACCTACTGTACCTACGCC Ⅲ F3(1) F CGCGGATCCAATTTGGACAGGGCGGCGGCGGTGCGCTTCATGTCGAATATCATTTATATAC Ⅲ F3(2) F CGCGGATCCGACAGGGCGGCGGCGGTGCGCGACGGTTCATGTCGAATATCATTTATATAC Ⅲ F3(3) F CGCGGATCCGCGGCGGCGGTGCGCGACGGTGCCGAATTCATGTCGAATATCATTTATATAC Ⅲ F3(4) F CGCGGATCCGCGGTGCGCGACGGTGCCGAAAATGAATTCATGTCGAATATCATTTATATAC Ⅲ F3(5) F CGCGGATCCCGCGACGGTGCCGAAAATGAAATACAAACGTTCATGTCGAATATCATTTATATAC Ⅳ F4(1) F CGCGGATCCACAAACGATACGGATCAACTTTTTCCCTTCATGTCGAATATCATTTATATAC Ⅳ F4(2) F CGCGGATCCGATACGGATCAACTTTTTCCCGAAAGATTCATGTCGAATATCATTTATATAC Ⅳ F4(3) F CGCGGATCCGATCAACTTTTTCCCGAAAGATTCGAATTCATGTCGAATATCATTTATATAC Ⅳ F4(4) F CGCGGATCCCTTTTTCCCGAAAGATTCGAAGGTTTTTTCATGTCGAATATCATTTATATAC Ⅳ F4(5) F CGCGGATCCCCCGAAAGATTCGAAGGTTTTTTCAACTTCATGTCGAATATCATTTATATAC Ⅳ F4(6) F CGCGGATCCAGATTCGAAGGTTTTTTCAACGAAGCCTTCATGTCGAATATCATTTATATAC Ⅳ F4(7) F CGCGGATCCGAAGGTTTTTTCAACGAAGCTATTGCTTCATGTCGAATATCATTTATATAC a The reverse primer used for the amplification of fragments Ⅲ F1 - Ⅳ F4(7) was the same as that used for the amplification of fragment Ⅰ; b Primer sequences are 5′ to 3′; c Restriction enzyme sites are underlined; d F indicates forward primer; R indicates reverse primer. Table 1. Primers for the PCR amplification of truncated and chimeric P74 gene fragments
-
Monoclonal antibodies against HearNPV P74 were prepared as reported in the literature (Evan G I et al., 1985). BALB/c mice were immunized with purified HearNPV P74△3TM. Spleen cells were then harvested from the immunized mice and fused with SP2/0 myeloma cells. Cell culture supernatants were screened by Western blotting, using HearNPV ODV (data not shown). Positive hybridomas were selected and further cloned and then inoculated into syngeneic mice, following the protocol approved by the Animal Ethics Committee at the Wuhan Institute of Virology, Chinese Academy of Sciences (CAS). Ascites fluids from three clones were withdrawn and clarified, and designated as 20D9, 20F9 and 21E1.
The mAbs were purified from collected ascites fluids using a Protein A-SepharoseTM column. Ascites fluids were each diluted with an equal volume of phosphate-buffered saline (PBS), pH 8.0; each was then applied to the column and the column was then in each case washed with 10 column volumes of PBS (pH 8.0); the antibodies were then eluted with 100 mmol/L citric acid buffer (8.61 g citrate and 2.64 g citric acid sodium in 500 mL H2O, pH 3.0). Each eluate was mixed with 2/5 elution volume of 1 mol/L Tris/HCl buffer, pH 8.0. The concentrations of the resulting purified mAb preparations were adjusted to 1.5 mg/mL.
-
Purified prokaryotic expressed proteins were separated by 12% SDS-PAGE, and transferred onto a polyvinylidene fluoride (PVDF) membrane (Merck Millipore, Germany). The anti-P74 polyclonal antibody (Song J J et al., 2008) from the supernatant solution of P74 hybridoma or ascites fluids was used as the primary antibody and alkaline phosphatase (AP)-conjugated anti-mouse immunoglobulin G(Protein Tec Group, USA) was used as the secondary antibody. Signals were detected as previously described (Deng F et al., 2007).
-
Hz-AM1 cells (3×105) were infected with vHaBacHZ8-egfp-ph at a multiplicity of infection (MOI) of 10. At 48 h postinfection (p.i.), the HzAM1 cells, on 10 mm glass plates, were washed three times with PBS, fixed with 4 % (w/v) paraformaldehyde and then permeabilized with fresh 0.15 %(w/v)Triton X-100 (in PBS). The cells were then incubated with primary antibodies (anti-P74 polyclonal antibody, supernatant solution from P74 hybridoma or ascites fluids, or purified mAbs) at 4 ℃ overnight. DyLightTM 594 or Rhodamine-conjugated anti-mouse IgG (Jackson ImmunoResearch, USA) was then added and the cells were incubated at 25 ℃ for 2 h, before being washed three times with PBS. Hoechst 33258 (Sigma, USA) was used to stain the nucleus for 15 min at 25 ℃. After washing with PBS, the cells were examined using a Revolution XD confocal microscope (Andor Technology, UK).
Virus, cells, strains and plasmids
Construction and expression of recombinant plasmids
Production and purification of antibodies
Western blot analyses
Immunofluorescence microscopy
-
Western blot analyses were carried out using purified, prokaryotically expressed N, M and C as antigens. A 45 kDa band corresponding to the N fragment was detected when 20D9 and 21E1 (Fig. 2, lanes 1 and 2), but not 20F9 (Fig. 2, lane 3), were used as primary antibodies. No signals were detected with 20D9, 21E1 or 20F9 when M or C were used as antigens (Fig. 2, lanes 6-8 and 11-13). Thus, 20D9 and 21E1 each showed a specific reaction with N, whilst 20F9 was not capable of recognizing prokaryotically expressed P74 segments.

Figure 2. Binding of mAbs 20D9 and 21E1, but not 20F9, to the P74 N fragment. Purified N (lane 1-5), M (lane 6-10) and C (lane 11-15) fragments were separated by 12 % SDS-PAGE (0.08 μg/lane), and Western blot analyses were performed with 20D9, 21E1 and 20F9. Anti-P74 polyclonal antibodies (P) and pre-immune serum (S) were used as positive and negative controls, respectively. 'Marker' indicates molecular mass markers.
-
Using immunofluorescence microscopy, a similar localization pattern for P74 was demonstrated, whether using mAbs 20D9 or 20F9, or polyclonal antibody against P74. In each case, P74 was detected in the ring-zone region of vHaBacHz8-egfp-ph-infected cells (Fig. 3), in agreement with a previous study of AcMNPV P74 (Slack J M et al., 2001). However, no fluorescence was detected using 21E1, indicating that whereas both 20D9 and 20F9 can detect native P74, 21E1 cannot.

Figure 3. Recognition of P74 by mAbs 20D9 and 20F9 in HearNPV-infected cells. HzAM1 cells were infected with vHaBac-egfp-ph and, at 48 h p.i., cells were fixed in 4 % (w/v) formaldehyde. Immunofluorescence staining to detect HearNPV P74 (yellow) was performed using anti-P74 monoclonal antibodies and DyLightTM594-conjugated goat anti-mouse IgG. Hoechst 33258 was used for staining of the nucleus. PC, anti-P74 polyclonal antibodies; NC, pre-immune serum. Scale bar = 10 μm.
-
To screen therecognition regions of 20D9 and 21E1, the N fragment was sequentially truncated. In the first round of screening, Western blot analyses were conducted using purified prokaryotically expressed Ⅰ-Ⅳ as antigens. The results showed that 20D9 recognized fragment Ⅲ but not Ⅰ, Ⅱ, or Ⅳ (Fig. 4A), whilst 21E1 recognized fragment Ⅳ but not fragments Ⅰ-Ⅲ (Fig. 4B, lane 5).
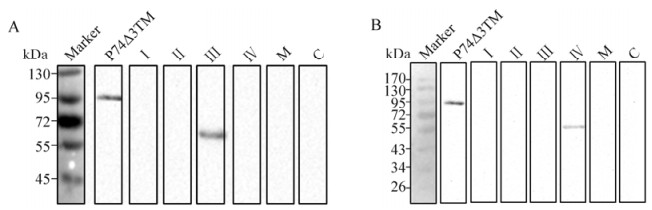
Figure 4. Localization of the recognition regions of 20D9 and 21E1 on fragments Ⅲ and Ⅳ, respectively, of N. Purified P74△3TM, Ⅰ, Ⅱ, Ⅲ, Ⅳ, M and C were separated by 12 % SDS-PAGE (0.08 μg/lane). Western blot analyses were performed with 20D9 (A) and 21E1 (B), using anti-P74 polyclonal antibodies (P) and pre-immune serum (S) as positive and negative controls, respectively. 'Marker' indicates molecular mass markers.
In the second round, Western blot assays were carried out using prokaryotically expressed chimeric P74 proteins as antigens. A 60 kDa band representing the chimeric protein Ⅲ F3 was detected by 20D9 (Fig. 5A, lane 3), whilst the chimeric protein Ⅳ F4 was detected by 21E1 (Fig. 5B, lane 4). Therefore, the recognition regions of 20D9 and 21E1 could be mapped to N136-T153 and T199-C219, respectively.
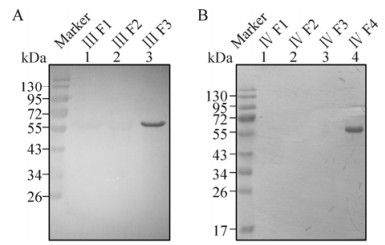
Figure 5. Mapping the recognition regions of 20D9 and 21E1. Purified chimeric proteins containing Ⅲ F1-F3 and Ⅳ F1-F4 were separated by 12 % SDS-PAGE (0.08 μg/lane). Western blot analyses were performed with 20D9 (A) and 21E1 (B), using anti-P74 polyclonal antibodies (P) and pre-immune serum (S) as positive and negative controls, respectively. 'Marker' indicates molecular mass markers.
The regions N136-T153 and T199-C219 were further truncated into five and seven peptides respectively, with an overlap of six amino acids between each pair of adjacent peptides (Fig. 1). A specific cross-reaction of 20D9 with Ⅲ F3(5) was observed (Fig. 6, lane 5) and thereby pinpointed the epitope recognized by 20D9 to R144-T153. However, no signal was detected when 21E1 was used as the primary antibody (data not shown).

Figure 6. Determination of the epitopes recognized by 20D9 and 21E1. Purified chimeric proteins containing Ⅲ F3(1)-(5) and Ⅳ F4(1)-(7) were separated by 12 % SDS-PAGE (0.08 μg/lane), and Western blot analyses were performed with 20D9 (A) and 21E1 (B). Anti-P74 polyclonal antibodies (P) and pre-immune serum (S) were used as positive and negative controls, respectively. 'Marker' indicates molecular mass markers.
-
Since 20D9 was capable of recognizing both prokaryotically and eukaryotically expressed P74, a purification procedure was undertaken. Western blot assays revealed that a signal representing P74△3TM could be detected using purified 20D9 mAbs, even at an antibody dilution of 1:20, 000 (Fig. 7A). Using purified 20D9 diluted × 500, a ring-zone location of P74 in vHaBac-egfp-ph-infected cells was observed under immunofluorescence microscopy (Fig. 7B), in agreement with work reported previously (Slack J M et al., 2001).
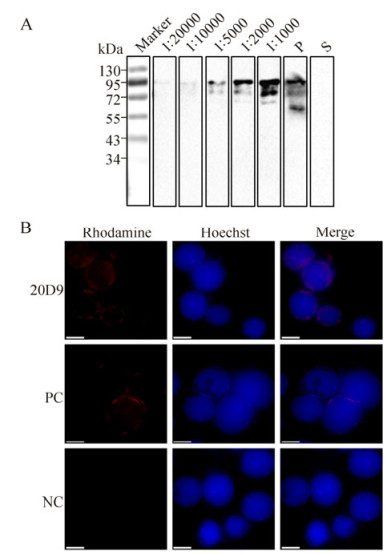
Figure 7. Characterization of purified 20D9. A: Purified P74△3TM was analyzed by 12 % SDS-PAGE (0.08 μg/lane), and Western blot analyses were performed with purified 20D9 diluted x 1, 000; x 2, 000; x 5, 000; x 10, 000 and x 20, 000, respectively (reading from right to left). Anti-P74 polyclonal antibodies (P) and pre-immune serum (S) were used as positive and negative controls, respectively. 'Marker' indicates molecular mass markers. B: HzAM1 cells were infected with vHaBac-egfp-ph, and at 48 h p.i., cells were fixed in 4% (w/v) formaldehyde. Immunofluorescence staining was performed using purified 20D9 at a dilution of 1:500 and Rhodamine-conjugated goat anti-mouse IgG to detect HearNPV P74 (red). Hoechst 33258 was used for nucleus staining. PC, polyclonal antibodies; NC, pre-immune serum. Scale bar=10 μm.
20D9 and 21E1, but not 20F9, bound specifically to the N fragment of P74
20D9 and 20F9 recognized native expressed P74 during HearNPV infection
Identification of the epitope regions recognized by 20D9 and 21E1
Purified 20D9 was of high titer and capable of recognizing both prokaryotically and eukaryotically expressed P74
-
In this study, we generated and characterized three mAbs against HearNPV P74, an important protein for baculovirus oral infection. In accordance with the putative cleavage sites (Peng K et al., 2011; Slack J M et al., 2008) (Fig. 1), P74 was first dissected into three fragments, which were then used for an initial round of screening. Two (20D9 and 20F9) out of the three mAbs clearly showed specific binding to the N fragment (Fig. 2). The N fragment was then further truncated to produce smaller fragments, and we were able to map the epitope regions of 20D9 and 20F9 to the sequences R144-T153 and T199-C219, respectively.
Normally, the smallest epitopes that can still be recognized by antibodies can be mapped to regions that are about six amino acids (aa) long (Abbas A K et al., 2012). However, binding between antibody and antigen would be optimized if additional amino acids were present on either side of the recognition region (Davies D R et al., 1990). During our epitope mapping, we found that 20D9, which bound to R144-T153, did not successfully recognize either Ⅲ F3(4), comprising aa 142-150, or fragment Ⅳ (aa 146-219). Similarly, although 20F9 bound to Ⅳ F4, it failed to react with any truncated Ⅳ F4 fragments [Ⅳ F4(1)-(7)] of length 9 aa. We attempted to use synthetic peptides to map the epitopes, but these attempts failed (data not shown). At this stage, therefore, we can only map 20D9 and 20F9 to epitopes of lengths of 10 and 20 aa, respectively. In the present study, to enable the extensive mapping of the epitopes, all truncated fragments smaller than 50 aa in length were fused with fragment Ⅰ (which has been shown no reaction with the mAbs) at their C terminus, for protein expression. This method is efficient for small fragment cloning and epitope mapping, as shown in Figs. 5 and 6.
The ability of 20D9 to recognize native P74 in HearNPV-infected cells (Figs. 3 and 7b) suggested that the R144-T153 region may naturally be exposed on the surface of HearNPV P74. Fragment 21E1 could also bind to native P74 (Fig. 3), although in this case no linear epitope was identified using the mAb, suggesting the recognition of a conformational epitope instead.AcMNPV P74 has been revealed to be loosely associated with a stable protein complex comprising PIF1, PIF2, PIF3 and PIF4 (Peng K et al., 2010; Peng K et al., 2012). The specific involvement of multiple, interacting PIFs in oral infection is indicative of the complexity of the oral infection process. The availability of 20D9 and 21E1 should facilitate the study of P74 and of its interactions with other PIFs during ODV infection.
In summary, we have characterized three mAbs of P74. One of these, 20D9, can detect a linear epitope of R144-T153 which is likely to be exposed on the surface of native P74. The second, 20F9, recognizes a linear epitope at T199-C219, whilst the third, 21E1, recognizes a conformational epitope. These mAbs will help us to understand the role of P74 during oral infection, especially its detailed cleavage and activation process.
-
This project was supported by the National Science Foundation of China (31130058 to Z.H.).
-
ZT, HW, FD and ZH designed the research; LL, TZ, HH and MW performed the experiment; DH, LL, ZH and TZ wrote the paper. The authors declare no conflict of interest.














 DownLoad:
DownLoad: