HTML
-
Baculoviridae, a family of enveloped rod-shaped viruses with single, circular double-stranded DNA genomes, is divided into four genera, i.e., Alphabaculovirus, Betabaculovirus, Gammabaculovirus, and Deltabaculovirus (King et al., 2012). Both alphabaculoviruses and betaba-culoviruses infect insects of the order Lepidoptera, while gamma-and deltabaculoviruses are found only in hymenopteran and dipteran insects, respectively. Alpha-, gamma-, and deltabaculoviruses produce crystalline occlusion bodies (OBs, or polyhedra) containing multiple virions with multiple or single nucleocapsids in the nuclei of host cells. Betabaculoviruses form OBs in the nuclei or cytoplasm of host cells. Each OB (granule) has a single virion with only one embedded nucleocapsid. Molecular analyses of baculoviruses have mainly focused on alphabaculoviruses, since permissive cell lines supporting efficient virus replication are limited to viruses in this genus.
In alphabaculoviruses, genes encoding proteins involved in early events in virus replication, such as gene expression and DNA replication, are typically transcribed by the host RNA polymerase Ⅱ, whereas genes encoding structural proteins and proteins associated with the assembly of progeny virions are transcribed by a virus-encoded RNA polymerase (Huh and Weaver, 1990; Hoopes and Rohrmann, 1991; Guarino et al., 1998). Viral gene transcription with the host RNA polymerase Ⅱ is initiated earlier than transcription catalyzed by the viral RNA polymerase. Accordingly, genes transcribed by the host RNA polymerase and the viral RNA polymerase are referred to as early and late genes, respectively. Some viral genes are transcribed by both the host and the viral RNA polymerases from different transcription initiation sites in promoter regions (Chen et al., 2013).
Late gene transcription is always initiated within the conserved late promoter motif T/G/ATAAG (Rohrmann, 1986; Rankin et al., 1988). In a transcriptome analysis of Autographa californica multiple nucleopolyhedrovirus (AcMNPV), the type species of the alphabaculoviruses, in a Trichoplusia ni cell line, 218 transcription initiation sites were identified in the viral genome. One hundred and twenty-six viral transcripts are initiated within the late promoter motif (Chen et al., 2013). Late gene expression relies on early gene expression and is also dependent on viral DNA replication (Rice and Miller, 1986). Based on transient expression assays, about 19 early genes are known to be required for late gene expression in AcMNPV (Rapp et al., 1998). These late expression factor (lef) genes are all early genes, although some are also transcribed by the viral RNA polymerase (Chen et al., 2013). Nine lef genes are involved in viral DNA replication (Kool et al., 1994; Lu and Miller, 1995). An additional nine or ten lef genes encode subunits of the viral RNA polymerase or other regulatory proteins, respectively (Guarino et al., 1998; Hayakawa et al., 2000).
Betabaculoviruses are the second largest group of baculoviruses. The genomes of 17 betabaculoviruses have been completely sequenced. However, information on the molecular mechanisms underlying replication in betabaculoviruses is very limited owing to the unavailability of efficient permissive cell lines. Based on published genome sequence data, betabaculovirus genomes have homologs of the genes encoding the RNA polymerase in alphabaculoviruses; additionally, the T/G/ATAAG motif, which is the core sequence of alphabaculovirus late gene promoters, is also present in the homologous genes of betabaculoviruses (Akiyoshi et al., 1985; Chakerian et al., 1985; King et al., 2005). However, it is still unclear whether late gene expression is regulated by similar mechanisms in betabaculoviruses and alphabaculoviruses. In a previous study, we constructed several reporter bacmids of the betabaculovirus Plutella xylostella granulovirus (PlxyGV) and used them to transfect three insect cell lines; we found that the promoters of the late genes vp39 and gran in the PlxyGV bacmids were not activated efficiently in transfected cells (Hu and Li, 2014). In the current study, we investigated if the late gene promoters of PlxyGV could be activated efficiently by the RNA polymerase of the alphabaculovirus AcMNPV.
-
The bacmid vPxG was derived from PlxyGV (Hu and Li, 2014). The bacmid bMON14272 was derived from the AcMNPV strain E2 (Luckow et al., 1993). They were separately maintained in DH10B cells as described previously (Luckow et al., 1993; Hu and Li, 2014; Huang et al., 2015).
The Sf9 cell line is a clonal isolate of the parent cell line IPLB-Sf21-AE derived from the fall armyworm Spodoptera frugiperda (Vaughn et al., 1977). The cells were cultured at 26℃ in Grace's medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum, penicillin (100 μg/mL), and streptomycin (100 μg/mL).
The DNA primers used in this study were synthesized by GenScript, Inc. (Piscataway, NJ, USA) and are shown in Supplementary Table S1.
-
Using PlxyGV DNA as a template, the putative promoter sequences of vp39, p6.9, e25, e18, gp41, pk1, polh, p10a, and p10b were PCR-amplified with the correspon-ding primer pairs Ppxvp39-UP and Ppxvp39-DP, Ppxvp39L-UP and Ppxvp39L-DP, Ppxp6.9-UP and Ppxp6.9-DP, Ppxe25-UP and Ppxe25-DP, Ppxe18-UP and Ppxe18-DP, Ppxgp41-UP and Ppxgp41-DP, Ppxgp41L-UP and Ppxgp41-DP, Ppxpk1-UP and Ppxpk1-DP, Ppxgran-UP and Ppxgran-DP, Ppxorf21-UP and Ppxorf21-DP, and Ppxorf50-UP and Ppxorf50-DP, described in Supplementary Table S1. The PCR products were digested with SnaB Ⅰ and BamH Ⅰ for Ppxvp39 and Ppxgp41L, or digested with BamH Ⅰ for Ppxp6.9, Ppxe25, Ppxe18, Ppxgp41, Ppxpk1, Ppxgran, Ppxorf21, and Ppxorf50, and inserted between the SnaB Ⅰ and BamH Ⅰ sites of pFB-luc (Hu and Li, 2014) to replace the polh promoter and make pPpxvp39-luc, pPpxgp41L-luc, pPpxp6.9-luc, pPpxe25-luc, pPpxe18-luc, pPpxgp41-luc, pPpxpk1-luc, pPpxgran-luc, pPpxp10b-luc, and pPpxp10c-luc. The PCR product of Ppxvp39L was digested with EcoR Ⅰ and inserted between the SnaB Ⅰ and EcoR Ⅰ sites of pFB-luc to construct pPpxvp39L-luc. Similarly, the promoter sequences of vp39, p6.9, e25, e18, gp41, and pk1 were PCR-amplified from AcMNPV DNA using the primer pairs Pacvp39-UP and Pacvp39-DP, Pacp6.9-UP and Pacp6.9-DP, Pace25-UP and Pace25-DP, Pace18-UP and Pace18-DP, Pacgp41-UP and Pacgp41-DP, and Pacpk1-UP and Pacpk1-DP. The PCR products were digested with SnaB Ⅰ and BamH Ⅰ for Pacvp39, Pacgp41, and Pacpk1, digested with Stu Ⅰ and BamH Ⅰ for Pace25, digested with Hinc Ⅱ and BamH Ⅰ for Pacp6.9, or digested with BamH Ⅰ for Pace18, and ligated with pFB-luc cut with SnaB Ⅰ and BamH Ⅰ, separately, to make pPacvp39-luc, pPacgp41-luc, pPacpk1-luc, pPace25-luc, pPacp6.9-luc, pPace18-luc, and pPacp10-luc (Figure 1).
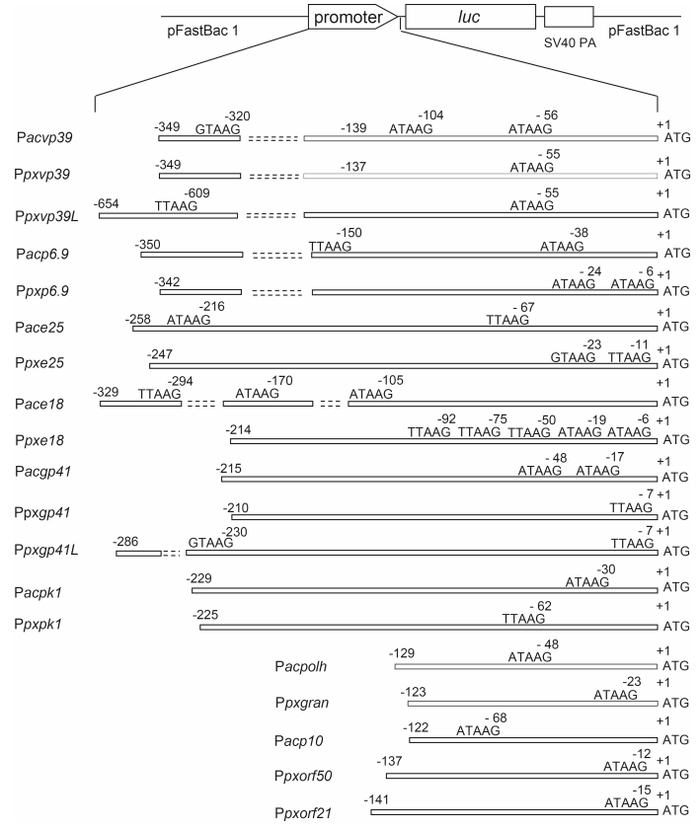
Figure 1. Structures of the reporter plasmids of nine/eight PlxyGV/AcMNPV late gene homologs. The late promoter motifs present in individual promoters and their locations relative to the translation initiation codon of each gene are indicated.
The reporter plasmids were separately transformed into Escherichia coli DH10B containing bMON14272 and the helper plasmid pMON7124 (Luckow et al., 1993), which encodes a transposase, to generate vAcPpxvp39-luc, vAcPpxvp39L-luc, vAcPpxvp6.9-luc, vAcPpxe25-luc, vAcPpxe18-luc, vAcPpxgp41-luc, vAcPpxgp41L-luc, vAcPpxpk1-luc, vAcPpxpgran-luc, vAcPpxp10b-luc, vAcPpxp10c-luc, vAcPacvp39-luc, vAcPacp6.9-luc, vAcPace25-luc, vAcPace18-luc, vAcPacgp41-luc, vAcPacpk1-luc, vAcPacpolh-luc, and vAcPacp10-luc.
Mutants of the PlxyGV e18 promoter were generated in which one or two of the TAAG motif(s) were mutated using the Fast Mutagenesis System (TransGene Biotech, Beijin, China), following the manufacturer's instructions. Briefly, the primer pairs Ppxe18-n7x-UP and Ppxe18-n7x-DP, Ppxe18-n20x-UP and Ppxe18-n20x-DP, Ppxe18-n76x-UP and Ppxe18-n76x-DP, Ppxe18-n95x-UP and Ppxe18-n95x-DP, were separately used for PCR with pPpxe18-luc as a template to construct pPpxe18n7x-luc, pPpxe18n20x-luc, pPpxe18n76x-luc, and pPpxe18n95x-luc. The primer pair Ppxe18-n20x-UP and Ppxe18-n20x-DP was used for PCR with pPpxe18n95x-luc as a template to make pPpxe18n20x95x-luc. The plasmids were separately transformed into E. coli DH10B containing bMON14272 and pMON7124 to generate vAcPpxe18n7x-luc, vAcPpxe18n20x-luc, vAcPpxe18n76x-luc, vAcPpxe18n95x-luc, and vAcPpxe18n20x95x-luc, respectively.
-
Transfection was performed as previously described (Hu and Li, 2014). Briefly, log-phase Sf9 cells were seeded in wells of a 96-well tissue culture plate at a density of 1 × 105 cells/well and incubated at 26℃ overnight. Following three washes with Grace's medium, the transfection mixture was added to the surface of the cell layer at 100 μL/well. The transfection mixture was made by mixing equal volumes of plasmid or bacmid (0.5 μg/50 μL in Grace's medium) and Lipofectin (1:50 diluted in Grace's medium). For mock transfection, no plasmid or bacmid was included in the transfection mixture. After incubation at 26℃ for 6 h, the transfection mixture was removed and replaced with fresh growth medium. For infection, the cells seeded in each well of a 96-well tissue culture plate were inoculated with 100 μL of 1:10 diluted infectious supernatant from transfected cells (MOI > 10) and incubated for 1 h. The inoculum was then removed and replaced with 100 μL of fresh growth medium. At 48 h post-transfection (hpt) or designated time points post-infection, the medium was removed, and the cells were lysed by the addition of lysis buffer (Promega, Madison, USA), following three washes with phosphate-buffered saline. Luciferase activity was determined using Luciferase Activity Reagent (Promega), and the chemiluminescent reactions were measured using a CentroXS3 LB960 (Berthold Technologies, Zug, Switzer-land). All luciferase expression values were derived from three independent transfections or infections for each construct set in each experiment.
Virus, cell line, and primers
Construction of reporter plasmids and bacmids
Transfection/infection of Sf9 cells and luciferase assay
-
The promoters of nine putative late genes in the PlxyGV genome were analyzed, including px79 (vp39), px67 (p6.9), px74 (odv-e25; e25), px13 (odv-e18, e18), px87 (gp41), px6 (pk1), px4 (gran), px21 (p10b), and px50 (p10c). Despite the lack of functional or expression data for these PlxyGV genes, their homologs in AcMNPV are typical late genes and play important roles in virus replication. In AcMNPV and other alphabaculoviruses, vp39 encodes the major capsid protein (Thiem and Miller, 1989). AcMNPV vp39 contains two ATAAG motifs at positions -56 and -104 and GTAAG at -320 upstream of the translation initiation codon ATG. PlxyGV vp39 has only one ATAAG at -55 and a TTAAG at position -609.
AcMNPV p6.9 encodes the sole viral DNA-binding protein, which is one of the three most abundant budded virus proteins (Wilson et al., 1987; Wang et al., 2010). There is an ATAAG at position -38 and a TTAAG at position -150 upstream of the AcMNPV p6.9 open reading frame. PlxyGV p6.9 has two ATAAG motifs in close proximity at positions -6 and -24.
The products of AcMNPV e25 and e18 are envelope proteins present in both budded viruses and occlusion-derived viruses (Russell and Rohrmann, 1993; Braunagel et al., 1996; 2003; Wang et al., 2010). There is a TTAAG at position -67 and an ATAAG at -216 in AcMNPV e25. The TTAAG motif is the start site for transcription (Chen et al., 2013). PlxyGV e25 has a TTAAG at position -11 and a GTAAG at -23. Two ATAAGs and one TTAAG are located at -105, -170, and -294, respectively, in the AcMNPV e18 promoter. In PlxyGV e18, two copies of ATAAG and three copies of TTAAG are located at positions -6, -19, -50, -75, and -92.
AcMNPV gp41 encodes a tegument glycoprotein localized between the virion envelope and the capsid in occlusion-derived viruses (Whitford and Faulkner, 1992a). Two copies of ATAAG at positions -17 and -48 serve as transcription start sites (Whitford and Faulkner, 1992b; Chen et al., 2013). PlxyGV gp41 has a TTAAG motif at position -7 and a GTAAG at -230. The pk1 gene encodes a protein kinase and is involved in very late gene expression (Reilly and Guarino, 1994; Mishra et al., 2008b). AcMNPV pk1 is transcribed from the sole ATAAG motif located at the -30 position (Reilly and Guarino, 1994; Chen et al., 2013). PlxyGV pk1 also has a single TTAAG at the -67 position.
polh and p10 are two very late genes that are expressed at high levels in the late and very late phases of infection in alphabaculoviruses. AcMNPV polh encodes the OB protein polyhedrin and has a single ATAAG motif at the -84 position, serving as the transcription start site (Russell et al., 1991; van Oers and Vlak, 1997). PlxyGV granulin (gran), which is homologous to AcMNPV polh, also has only one ATAAG at position -23. Similarly, AcMNPV p10 has a single ATAAG motif located at -68, functioning as the start site for transcription (Kuzio et al., 1984; Chen et al., 2013). In PlxyGV, there are three open reading frames that are homologous to AcMNPV p10, i.e., orf2 (p10a), orf21 (p10b), and orf50 (p10c). Since there is no late promoter motif within the immediate upstream region of orf2, only the promoters of orf21 and orf50 were included in this study.
The lengths of the cloned promoters and the locations of all T/G/ATAAG motifs within individual promoters are shown in Figure 1. No transcriptional initiation at any site other than the T/G/ATAAG motifs was detected for these AcMNPV late genes.
-
To examine the activities of the PlxyGV late gene promoters, reporter plasmids named pPpxvp39-luc, pPpxp6.9-luc, pPpxe25-luc, pPpxe18-luc, pPpxgp41-luc, pPpxpk1-luc, pPpxgran-luc, pPpxp10b-luc, and pPpxp10c-luc were constructed and used in transient assays. These reporter plasmids contained the coding sequence of the Photinus pyralis luciferase gene (luc) under the control of the individual PlxyGV late gene promoters (Figure 1). In transient assays, Sf9 cells were transfected with the individual reporter plasmids alone or together with the PlxyGV bacmid vPxG or AcMNPV bacmid bMON14272. The Sf9 cell line is permissive to AcMNPV and non-permissive to PlxyGV. As shown in Figure 2, the levels of luciferase (LUC) activity at 48 hpt for all transfections with individual reporter plasmids alone or together with vPxG were similar to the level observed in the mock transfection. In contrast, LUC levels in the cells transfected with mixtures of reporter plasmids and bMON14272 increased by 100-to 10, 000-fold compared with that in cells with the reporter plasmids alone. These results demonstrated that the PlxyGV late gene promoters could not be activated by the RNA polymerase of Sf9 cells, even in the presence of the PlxyGV genome, but they could be activated effectively in the presence of the AcMNPV genome. These results confirmed those of our previous study, in which luc and another reporter gene, egfp, linked with the PlxyGV vp39 or gran promoter could not be activated in the presence of the PlxyGV bacmid but could be activated in the presence of the AcMNPV bacmid (Hu and Li, 2014).

Figure 2. Transient assays of reporter gene expression from the late promoters PlxyGV vp39, p6.9, e25, e18, gp41, pk1, gran, p10b, and p10c in Sf9 cells. The graphs show luciferase (LUC) activity (in relative light units, RLU) measured at 48 hpt in cells transfected with the individual reporter plasmids pPpxvp39-luc, pPpxp6.9-luc, pPpxe25-luc and pPpxe18-luc, pPpxgp41-luc, pPpxpk1-luc, pPpxgran-luc, pPpxp10b-luc, and pPpxp10c-luc alone (grey columns), together with vPxG (blue columns) or bMON14272 (red columns), or mock-transfected (black columns, no any reporter plasmid or bacmid included). The names of the reporter plasmids are placed below each group of columns. Error bars represent the standard error from three independent experiments.
-
Since the PlxyGV late gene promoters could be activated by the AcMNPV bacmid, additional transient assays were performed to compare the activities of the PlxyGV late gene promoters with those of the promoters of the corresponding AcMNPV homologs in the presence of the AcMNPV bacmid. The reporter plasmids pPacvp39-luc, pPacp6.9-luc, pPace25-luc, pPace18-luc, pPacgp41-luc, pPacpk1-luc, and pPacp10-luc, which contained the promoter sequences of AcMNPV vp39, p6.9, e25, e18, gp41, pk1, and p10, respectively, were constructed in the same way as the PlxyGV reporter plasmids and used in transient assays for comparative analyses. pFB-luc (named pPacpolh-luc in this study) containing luc under the control of the AcMNPV polh promoter (Hu and Li, 2014) was also included in the experiments. Considering that the length of the promoter may influence the expression level (Yang et al., 2014), the lengths of the AcMNPV late gene promoters contained in the reporter plasmids were made similar to those of the late gene promoters in the corresponding reporter plasmids of PlxyGV. The e18 promoter in pPpxe18-luc was 115 nt shorter than that in pPace18-luc since the TAAG motifs were much closer to the initiation codon ATG in PlxyGV e18 than in AcMNPV e18. Accordingly, two additional reporter plasmids, pPpxvp39L-luc and pPpxgp41L-luc, with extended promoter lengths were included in these transient assays. The PlxyGV vp39 promoter in pPpxvp39L-luc was 305 nt longer than that contained in pPpxvp39-luc with GTAAG at the -609 position. The gp41 promoter in pPpxgp41L-luc was 76 nt longer than that in pPpxgp41-luc and contained an additional GTAAG at the -230 position.
The individual reporter plasmids of PlxyGV and AcMNPV late gene promoters were mixed with bMON14272 and transfected into Sf9 cells. The LUC activity levels in cells with the reporter plasmids of PlxyGV vp39, p6.9, gp41, and pk1 were 16.2%/35.98%, 39.6%, 17.7%/30.2%, and 16.2% of the levels in cells with reporter plasmids of AcMNPV vp39, p6.9, gp41, and pk1, respectively. The levels in the cells transfected with the reporter plasmids of PlxyGV e25, e18, and gran were 6.2, 14.6, and 7.5 times greater than the levels observed in cells with the reporter plasmids of corresponding AcMNPV late genes. LUC activity levels in the cells with PlxyGV p10b and p10c were 118% and 87.1% of the levels in cells with the AcMNPV p10 reporter plasmid, respectively (Figure 3).
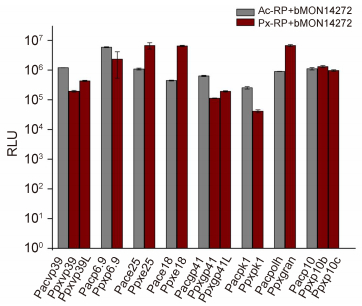
Figure 3. Comparative analysis of reporter gene expression from individual PlxyGV late promoters and corresponding AcMNPV late promoters by transient assays. The graphs show luciferase (LUC) activity (in relative light units, RLU) measured at 48 hpt in cells co-transfected with bMON14272 and the individual reporter plasmids of PlxyGV late promoters (red columns) or individual reporter plasmids of AcMNPV late promoters (grey columns). The names of the reporter plasmids are shown below each column. Error bars represent the standard error for three independent experiments.
-
To examine the activity levels of PlxyGV late promoters in the context of the AcMNPV genome in infection, eleven AcMNPV recombinant bacmids containing individual PlxyGV late gene promoters linked with luc were constructed, i.e., vAcPpxvp39-luc, vAcPpxvp39L-luc, vAcPpxvp6.9-luc, vAcPpxe25-luc, vAcPpxe18-luc, vAcPpxgp41-luc, vAcPpxgp41L-luc, vAcPpxpk1-luc, vAcPpxgran-luc, vAcPpxp10b-luc, and vAcPpxp10c-luc (Figure 4A). Similarly, eight recombinant bacmids containing the AcMNPV late gene promoters vAcPacvp39-luc, vAcPacp6.9-luc, vAcPace25-luc, vAcPace18-luc, vAcPacgp41-luc, vAcPacpk1-luc, vAcPacpolh-luc, and vAcPacp10-luc were generated for comparison (Figure 4A). The individual recombinant bacmids were transfected into Sf9 cells to produce reporter viruses. The infectious supernatants collected from the transfections were inoculated onto the surface of the Sf9 cell layer in the wells of 96-well plates. At the designated time points post-infection, the cells were sampled to measure luciferase activity.
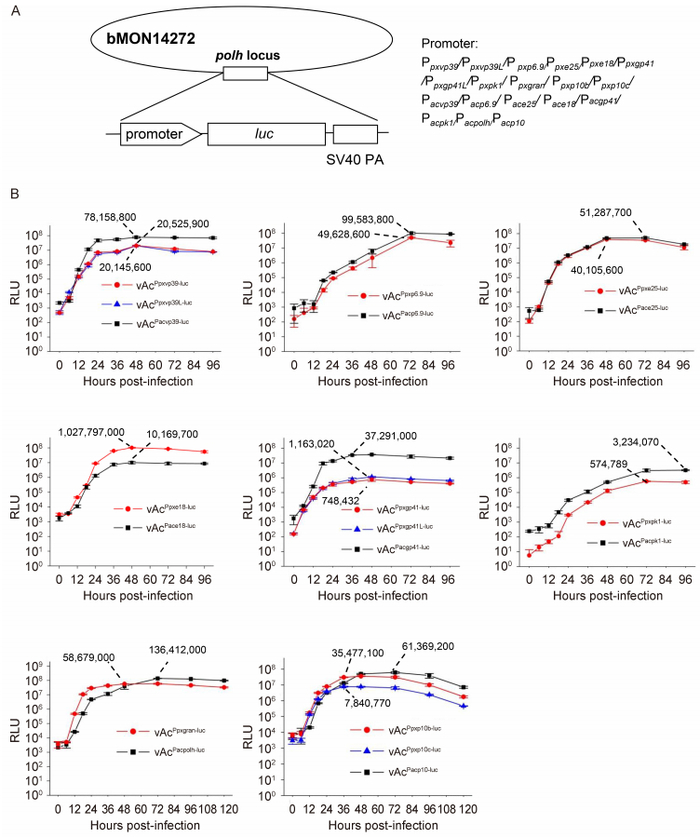
Figure 4. Comparative time-course analysis of reporter gene expression from individual PlxyGV late promoters and corresponding AcMNPV late promoters in infected cells. The graphs show luciferase (LUC) activity (in relative light units, RLU) measured at 0, 6, 12, 18, 24, 36, 48, 72, and 96 hpi for the promoters of vp39, p6.9, e25, e18, gp41, and pk1, or at 0, 6, 12, 18, 24, 36, 48, 72, 96, and 120 hpi for the promoters of polh/gran and p10/p10b/p10c, in cells infected by individual reporter viruses of PlxyGV late promoters (red or purple curves) or individual reporter plasmids of AcMNPV late promoters (black or gray curves). The numbers on the curves are the peak expression levels for each late promoter. Error bars represent the standard error for three independent experiments.
Figure 4B shows the time course of luciferase accumulation in Sf9 cells infected by individual reporter viruses of PlxyGV or AcMNPV late gene promoters, demonstrated by LUC activity levels in the cell extracts. In the cells infected by vAcPacvp39-luc, LUC activity increased dramatically between 6 and 24 hpi, continued increasing slowly, and reached a peak at 48 hpi. Thereafter, the LUC levels did not change significantly. In the cells infected by vAcPpxvp39-luc or vAcPPxvp39L-luc, LUC activity increased dramatically immediately after infection until 24 hpi, again reached peak levels at 48 hpi, and continued to remain high for the remainder of the study period. However, the acceleration in LUC activity in cells with vAcPpxvp39-luc or vAcPpxvp39L-luc was not as fast as that in the cells with vAcPacvp39-luc. The maximal LUC activity detected in the cells with vAcPpxvp39-luc was only 26.3% of the maximal level observed in cells with vAcPacvp39-luc. The lower activity observed for the PlxyGV vp39 promoter than the AcMNPV vp39 promoter may be due to the difference in the number of G/ATAAG motifs present in these two promoters. All three G/ATAAG motifs in AcMNPV vp39 are active start sites for transcription (Thiem and Miller, 1989; Chen et al., 2013). The activity levels detected in cells with vAcPpxvp39-luc were similar to those of cells with vAcPpxvp39L-luc at all time points, although the vp39 promoter in vAcPpxvp39L-luc had an extra TTAAG motif. This suggested that the TTAAG motif at -609 bp likely did not function as a transcription start site for PlxyGV vp39 in infection.
The accumulation of LUC activity in cells infected by both vAcPpxvp6.9-luc and vAcPacp6.9-luc demonstrated similar patterns, increasing steadily from 12 to 72 hpi. However, the levels in cells with vAcPpxvp6.9-luc were less than 50% of the LUC levels in cells with vAcPacp6.9-luc at most time points post-infection. AcMNPV p6.9 is transcribed solely from the ATAAG motif at the -38 position, although there is a TTAAG motif at the -150 position (Wilson et al., 1987; Chen et al., 2013). It is not clear whether both of the two closely positioned ATAAG motifs in the PlxyGV p6.9 promoter are active transcription start sites. Sequences around the ATAAG motif in the promoters may contribute to the difference in reporter gene expression between vAcPpxvp6.9-luc and vAcPacp6.9-luc in this case.
The levels of LUC activity detected in cells infected by both vAcPpxe25-luc and vAcPace25-luc were very similar at all time points. The ATAAG motif in the AcMNPV e25 promoter and the TTAAG motif in the PlxyGV e25 promoter and the difference in distance from the motifs to the initiation ATG did not cause a significant difference in reporter gene expression between these two e25 promoters.
An obvious difference in reporter gene expression was observed between vAcPpxe18-luc and vAcPace18-luc. In cells with vAcPpxe18-luc, LUC activity increased dramatically between 6 and 24 hpi. In cells infected by vAcPace18-luc, LUC activity increased immediately after infection, but the rate of acceleration was lower than that in cells with vAcPpxe18-luc. LUC levels in cells with vAcPace18-luc were much lower than those in cells infected with vAcPpxe18-luc at all time points after 12 hpi. The peak level in cells with vAcPpxe18-luc was 5.3 times higher than the peak levels in cells with vAcPace18-luc. In this case, the number of T/ATAAG motifs likely had a strong effect on reporter gene expression.
To verify the effects of individual T/ATAAG motifs on the promoter activity of PlxyGV, five reporter viruses containing luc under the control of a mutated PlxyGV promoter, in which one or two of the five motifs were mutated, were constructed by inserting the individual PlxyGV promoter-luc chimera to the polh locus of bMON14272 (Figure 5A). As shown in Figure 5B, LUC activity levels in the cells infected by vAcPpxe18n95x-luc were reduced obviously at all time points after 12 hpi in comparison with the activity levels in cells with vAcPpxe18-luc. However, the levels of LUC activity in the cells with vAcPpxe18n7x-luc, vAcPxpe18n20x-luc, or vAcPpxe18n76x-luc were similar or even slightly higher than those in the cells with vAcPpxe18-luc at all time points. In the cells infected with vAcPpxe18n20x95x-luc, which contained double point mutations within the ATAAG motif at -20 bp and the TTAAG motif at -96 bp, the LUC activity levels were lower than those in the cells with vAcPpxe18-luc, but were similar to those in cells with vAcPpxe18n95x-luc. LUC activity levels in cells with any one of the reporter viruses containing a mutated PlxyGV promoter were still much higher than those in cells with vAcPace18-luc.
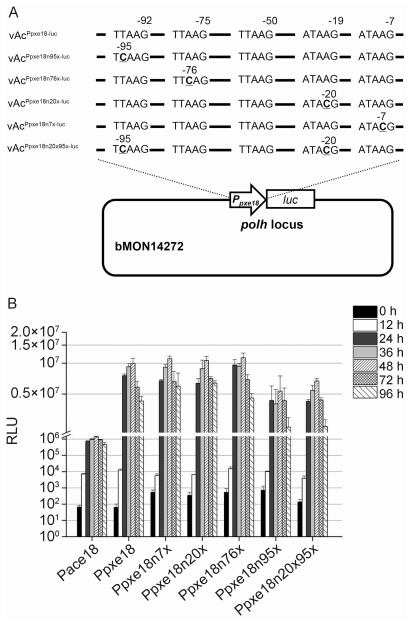
Figure 5. Effects of mutations in the late promoter motifs on the activity of the PlxyGV e18 promoter. (A) Construction of reporter viruses for mutants of the PlxyGV e18 promoter. The late promoter motifs present in the promoter and their locations relative to the translation initiation codon are indicated. The mutated nucleotides are presented in bold and underlined, and their locations are indicated. (B) Comparative analysis of reporter gene expression from individual mutants of the PlxyGV e18 promoter. The graphs show luciferase (LUC) activity (in relative light units, RLU) measured at 0, 12, 24, 36, 48, 72, and 96 hpi in the cells infected by individual reporter viruses as indicated. Error bars represent the standard error from three independent experiments.
In contrast to reporter gene expression from the e18 promoters, luc expression from the PlxyGV gp41 promoter was much less efficient than that from the AcMNPV gp41 promoter. LUC activity levels increased dramatically immediately after infection until 18 hpi in all cells infected by vAcPpxgp41-luc, vAcPpxgp41L-luc, or vAcPacgp41-luc, but the rate of acceleration was much higher in the cells with vAcPacgp41-luc than in the cells infected by vAcPpxgp41-luc or vAcPpxgp41L-luc. The maximal LUC activities detected in the cells with vAcPpxgp41-luc or vAcPpxgp41L-luc were only 2% and 3.2% of the levels observed in cells with vAcPacgp41-luc. The PlxyGV gp41 promoter sequence in vAcPpxgp41-luc had only one TTAAG motif, while AcMNPV gp41 had two closely located ATAAG motifs that have been identified as transcription start sites (Whitford and Faulkner, 1992b; Chen et al., 2013). The GTAAG at -229 bp in vAcPpxgp41L-luc may not be an efficient transcription start site.
Similar to the results of transient assays, the pk1 promoters of PlxyGV and AcMNPV were the weakest among the late gene promoters analyzed in infected cells. LUC activity levels increased steadily, with low rates of acceleration, throughout the whole process in cells infected by vAcPpxpk1-luc and cells with vAcPacpk1-luc. The levels detected in the cells with vAcPpxpk1-luc were lower than those in the cells with vAcPacpk1-luc at all time points. At 96 hpi, the levels of LUC accumulation in the cells with vAcPpxpk1-luc were only 17.8% of the levels observed in cells with vAcPacpk1-luc.
Reporter gene expression levels in cells infected by viruses containing the PlxyGV gran, p10b, or p10c promoters were higher at the early stage and lower at the late stage than in cells infected by the viruses containing the polh promoter or an additional copy of the p10 promoter. In alphabaculoviruses, both polh and p10 are associated with polyhedra formation. Polyhedra are much larger than granules and contain multiple enveloped nucleocapsids. Accordingly, a longer period and more polyhedrin proteins may be needed for the production of polyhedra than those required for the production of granules. The difference in the temporal expression of polh/gran and p10 between PlxyGV and AcMNPV may reflect the differential requirement for the proteins by these two viruses.
In cells infected by vAcPacpolh-luc, the levels of LUC activity continued to increase and reached a peak at 72 hpi, after which they did not change significantly, while the levels of LUC activity in cells with vAcPpxgran-luc did not increase obviously after 48 hpi. In cells infected by vAcPpxp10b-luc, vAcPpxp10c-luc, or vAcPacp10-luc, LUC activity levels reached a peak at 36 and 72 hpi, respectively. The peak levels in cells with vAcPpxp10b-luc and vAcPpxp10c-luc were 57.8% and 12.8% of the peak levels in cells with vAcPacp10-luc, respectively. In contrast to expression patterns for the polh/gran promoters, LUC activity levels decreased steadily in cells infected by the viruses in which luc was under the control of the p10 promoters after reaching peak levels. This might reflect competition from the expression of the native p10 gene. In viruses containing the reporter gene under the control of the polh or gran promoters, native polh was deleted, and reporter gene expression was not affected by competition from native gene expression.
Late promoter motifs of nine PlxyGV late genes and their AcMNPV homologs
PlxyGV late gene promoters were activated by an AcMNPV bacmid, but not by a PlxyGV bacmid in Sf9 cells
Activity of the late promoters of homologous PlxyGV and AcMNPV genes in the presence of the AcMNPV genome in transient assays
Time course of reporter gene expression from the PlxyGV late gene promoters integrated in the AcMNPV genome in infected cells
-
All nine late gene promoters of PlxyGV examined in this study were effectively activated in the context of the AcMNPV genome in a non-permissive cell line, indicating that PlxyGV late gene promoters could be recognized and transcribed by the RNA polymerase encoded by AcMNPV. These results suggest that the late gene expression machinery is conserved between alphabaculoviruses and betabaculoviruses. Baculoviruses, such as AcMNPV, are widely used as expression vectors for the production of various bioactive proteins and are used for the bio-control of crop pests. Late gene promoters with high activity levels from different viral species may be interchanged to enhance their efficiency via genetic manipulation.
Among the promoters of the nine PlxyGV and eight AcMNPV late gene homologs analyzed in this study, the numbers of G/T/ATAAG motifs present in the promoters of vp39, e18, and gp41 were different between PlxyGV and AcMNPV. In these cases, the number of promoter motifs was positively associated with reporter gene expression. In the promoter of AcMNPV e18, all three motifs upstream of the initiation codon ATG are start sites for transcription and are required for optimal virus replication (Braunagel et al., 1996; McCarthy and Theilmann, 2008; Chen et al., 2013). To verify whether or not the relative levels of reporter gene expression for the promoters of PlxyGV e18 and AcMNPV e18 were determined by the number of late promoter motifs, four of the five late promoter motifs of PlxyGV e18 were individually mutated and the activity levels of the mutant and native promoters were compared. The point mutation changing TTAAG at position -92 to TCAAG caused a significant reduction in reporter gene expression, whereas point mutations within the motifs at positions -6, -19, or -75 had little effect on promoter activity. It is unclear how the other nucleotides within each motif and the sequences flanking the individual motifs affect promoter activity; accordingly, additional experiments are necessary.
In the cases in which equal numbers of late promoter motifs were present in the promoters of two homologous genes, the expression levels for the AcMNPV promoters were always higher than those for the PlxyGV promoters, except for the e25 promoters, for which the levels of reporter gene expression were similar between the two homologs. These results implied that AcMNPV-specific elements contained in some of the AcMNPV late promoters were favored by the native viral RNA polymerase for optimal transcription. The promoters of polh, p10, and pk1 all contained a single T/ATAAG motif in both PlxyGV and AcMNPV. The peak levels of reporter gene expression using the promoters of PlxyGV gran, p10, and pk1 were lower than those for the promoters of the corresponding AcMNPV homologs (Figure 4). AcMNPV pk1 is expressed at the late and very late phases and is involved in the regulation of expression associated with the polh promoter (Reilly and Guarino, 1994; Fan et al., 1996; Mishra et al., 2008a; Mishra et al., 2008b). Alignments of the promoter sequences of PlxyGV and AcMNPV gran/polh, p10, and pk1 revealed an 8-nt sequence, TAAATAAG, encompassing the ATAAG motif that was conserved in the AcMNPV very late gene promoters and a 5-nt sequence, CAATT, located 4 or 5 nt upstream of the T/ATAAG motif in the promoters of PlxyGV gran, p10c, and pk1 (Figure 6). The effects of the TAA upstream of the ATAAG motif in the AcMNPV promoters and the CAATT sequence in the PlxyGV promoters on the temporal expression of viral genes remain to be determined.

Figure 6. Sequences conserved in very late gene promoters of PlxyGV and AcMNPV. Sequences from 10 nt downstream to 10 nt upstream of the transcription start sites of very late genes were compared for PlxyGV and AcMNPV. The 5-nt sequence CAATT conserved in the promoters of PlxyGV pk1, gran, and p10c and the 8-nt sequence TAAATAAG conserved in the AcMNPV pk1, polh, and p10 promoters are shown in bold. The transcription start site motifs are shown in bold and underlined.
-
This work was supported by the fund of Hubei Key Laboratory of Genetic Regulation and Integrative Biology.
-
The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
-
LLL, HLR, and YH designed the experiments. HLR, YH, and YJG carried out the experiments. LLL, HLR and YH analyzed the data. LLL wrote the paper. All authors read and approved the final manuscript.
Supplementary Table S1 is available on the website of Virologica Sinica: www.virosin.org; link.springer.com/journal/12250.
-
Name Sequence (5′-3′)a RE site Pacvp39-UP CGATTACGTACTCAAGTTGTGCGAAAC SnaB Ⅰ Pacvp39-DP CTCGGATCCAATATTGTTGCCGTTATAAAT BamH Ⅰ Ppxvp39-UP TGCATACGTAGGATCCCCGTAAAC SnaB Ⅰ Ppxvp39-DP GGCGGGATCCAATCATTTTAATGTTTTAATT BamH Ⅰ Ppxvp39L-UP TCATCGTGTGCGGTAAAAAGGGTTC - Ppxvp39-DP CCGGAATTCTCGCAGGCTAATAATTTTAATGTTT EcoR Ⅰ Pacp6.9-UP GGTCGACGTACCAAATTCCGTTTTGCGACG - Pacp6.9-DP GGTCGACGGATCCGTTTAAATTGTGTAATTTATG BamH Ⅰ Ppxp6.9-UP CCCCTCGTTATCGAAAACATTGCCAT - Ppxp6.9-DP ATAGGATCCGGCGTCTTACCAGTTTGTCT BamH Ⅰ Pace25-UP CTAAGGCCTTGTTCGATGCAATGAT Stu Ⅰ Pace25-DP CCCCACAGGATCCGTTTTAATTTAC BamH Ⅰ Ppxe25-UP CTGAATACGGTAGCTCGTCGGCG - Ppxe25-DP CGCGGATCCCAACAGTTTTTATTCTC BamH Ⅰ Pace18-UP CGCGCCCAAGCAGCGTATATTAAGT - Pace18-DP CGCGGATCCATTATTGTACCGAGTC BamH Ⅰ Ppxe18-UP CTCAAACCGGGAGACGTCCTGTACC - Ppxe18-DP CGCGCGGATCCCTTATTATTTTATCTTATG BamH Ⅰ Pacgp41-UP GCATTACGTATCGGCGCGTCAGTTTT SnaB Ⅰ Pacgp41-DP CGGGCATCTGGGATCCTTTTTATTTGT BamH Ⅰ Ppxgp41-UP TTGCGTGCTGCTAACCGTATTGAT - Ppxgp41-DP TCTGGATCCTAGTCTTAAAGATAACGGC BamH Ⅰ Ppxgp41L-UP GAGCTGACCGAATACGTAGAGGGAG SnaB Ⅰ Ppxgp41-DP TCTGGATCCTAGTCTTAAAGATAACGGC BamH Ⅰ Pacpk1-UP TCGTCGACGTACGTATATGCTTTGTTGT SnaB Ⅰ Pacpk1-DP TGGCGGATCCGAATCGTAGATATGAAT BamH Ⅰ Ppxpk1-UP AATCGACGTACGAGTGCGTGGAG - Ppxpk1-DP GCGTGGATCCAAACGATAACAATGT BamH Ⅰ Ppxgran-UP GCCCAACATACCGGACGCTATCACAAC - Ppxgran-DP CGCGGATCCAATGTTTTTGTAAAAATAAAAATTC BamH Ⅰ Ppxorf21-UP GGCGTTTCACGTAAAATTGTGCTCAT - Ppxorf21-DP TACGGATCCTGGAGATGTTATCGAAATT BamH Ⅰ Ppxorf50-UP TTTACGTACTGCGGAAACGGTTGC - Ppxorf50-DP GGCGGGATCCGTAAATCTTCTTATGTAT BamH Ⅰ Ppxe18n95x-U GATCGACAAGTAATGTCAAGGGTGTAAAAAAATT - Ppxe18n95x-D GACATTACTTGTCGATCGCACAATTATGTACAT - Ppxe18n76x-U GGGTGTAAAAAAATTACGTCAATTAAAAAAC - Ppxe18n76x-D GTAATTTTTTTACACCCTTAACATTACTTG - Ppxe18n20x-U TAAATTTATTTGTAACATACGATAAAATAATAAG - Ppxe18n20x-D GTATGTTACAAATAAATTTAAAAGCGTATTC - Ppxe18n7x-UP CATAAGATAAAATAATACGGGATCCCGGTC - Ppxe18n7x-DP GTATTATTTTATCTTATGTTACAAATAAATTT - Note: a sequences of restriction sites are underlined; the mutated late promoter motifs of PlxyGV e18 promoter are in bold and underlined. Table S1. PCR primers used in this study
















 DownLoad:
DownLoad: