-
Dear Editors,
The coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2, broke out in early December 2019 has escalated into a global pandemic (Lai et al. 2020). Till the May 20th 2020, more than 4, 700, 000 people were infected and the number is still increasing especially in Europe, North America and Asia (https://covid19.who.int/). And there is an urgent need to understand the biology of SARS-CoV-2. Here we report the isolation and characterization of seven isolates of SARS-CoV-2 from seven patients. All these isolates were adapted to grow in Vero cells, and showed significant different replication characteristics. Sequence analysis revealed that these viruses shared similar sequences to reported SARS-CoV-2 strains, except for 8#.
All of seven COVID-19 cases, from different cities of Zhejiang Province, were confirmed by real-time RT-PCR around 2020/01/26 (2020/01/24–2020/01/28). Basic information of those patients were listed in Table 1. Vero cells were seeded overnight in MEM containing 10% FBS before samples (processed swabs or sputum) were inoculated. After incubation for an hour, the cells were replenished with fresh maintain medium (MEM containing 3% FBS). The cells were observed daily for cytopathic effect (CPE) from the 2nd to the 7th days (Fig. 1A). At the day 7, the cell culture supernatant was collected for virus titration (Table 1).
Virus Gender Location Confirmed time Sample Ct value Clade Titer (TCID50/mL) 4# Female Shaoxing 2020/01/24 Pharyngeal swab 22 L 5.25 8# Male Hangzhou 2020/01/26 Pharyngeal swab 26 L 5.0 11# Male Wenzhou 2020/01/28 Sputum 22 S 5.25 12# Male Wenzhou 2020/01/26 Sputum 28 S 6.25 16# Male Ningbo 2020/01/24 Pharyngeal swab 23 S 5.0 17# Female Shaoxing 2020/01/27 Pharyngeal swab 26 S 3.25 19# Male Huzhou 2020/01/25 Pharyngeal swab 24 L 6.25 Table 1. Basic information of the isolated strains.
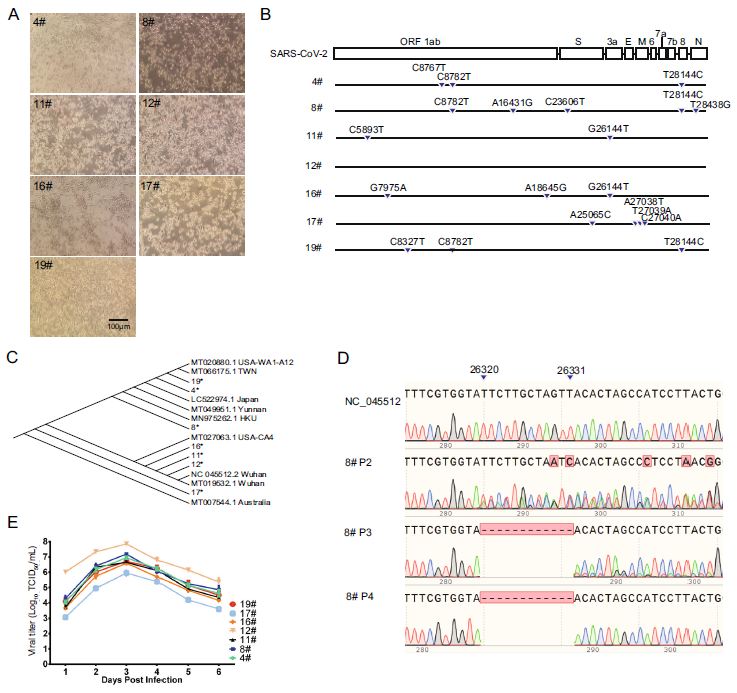
Figure 1. Characteristics of SARS-CoV-2. A The cytopathic effect was observed in Vero cells infected with the isolated viruses at 5 dpi. B Nucleotide mutations of isolated viruses at passage 3. The SARS-CoV-2 infected-cells were homogenized with Trizol, and total RNA was extracted and was used for RT-PCR as described below. First-strand cDNA was synthesized with oligo(dT) primers, then the complete genome was segmented amplified and sequenced. Sequencing results were analyzed by SnapGene. C Phylogenetic tree was generated by Clustal Omega based on amino acid sequence of seven isolates and part of the available viruses. D Sequence results of 8#, from Passage 2 to Passage 4. E The multiple growth curve of isolated viruses. The virus growth curves were drawn based on the viral titers measured at each indicated time point. The growth curves were obtained by three independent experiments.
It is well known that mutation on viral genes may have an impact on virulence, immunogenicity and other characterizations of viruses. In a recent report, 149 mutations were found among 103 sequenced isolates of SARS-CoV-2 (Tang et al. 2020). To investigate whether our viruses show mutations different from other reported SARS-CoV-2, all the seven isolates were sequenced at the 3rd passage and the sequences were aligned with coding sequence (CDS) of SARS-CoV-2 Wuhan-Hu-1 (NC_045512). Three and four of the seven isolates belong to the L and the S clades, respectively (Table 1) (Tang et al. 2020). It is interesting to note that these seven patients infected with SARS-CoV-2 have high viral load in the early stage of clinical sign, which is consistent with previous reports (Kim et al. 2020; Zou et al. 2020). As shown in Fig. 1B, genomic sequences of the viruses share sequence identity higher than 99.9%. The phylogenetic analysis of the isolated viruses together with other available viruses indicates that the seven isolates are closely related to each other, as well as to the other published sequences, which is consistent with the previous studies (Lu et al. 2020; Ren et al. 2020; Zhou et al. 2020) (Fig. 1C). And, each virus has 2–5 mutations except for isolate 12#, which is exactly the same as Wuhan-Hu-1. It's worth noting that apart from the 5 mutations, sequence of isolate 8# shows obvious gene deletion at 26320–26331 (E. gene) (Fig. 1D, 8# P3). To confirm this, isolate 8# at passage 2 and 4 was sequenced (Fig. 1D, 8# P2, 8# P4). Sequencing result showed that the nesting peak gradually weakened from P2 to P4 (Fig. 1D). At P4, a 12-base-deficient virus was predominant. The in vitro and in vivo effect of the deletion on this isolate should be further studied. On the contrary, isolate 12# did not mutate temporarily (P3) during in vitro passage. These results indicate that mutation occurred during virus passage in Vero cells, suggesting that virus purification and genetic monitoring are necessary in both vaccine development and etiology research.
To investigate the growth characteristics of seven isolated viruses, Vero cell was infected with each virus at a MOI of 0.001. The supernatant was collected for virus titration at 1, 2, 3, 4, 5, 6 days post infection (dpi). For the virus titration, Vero cells seeded in 96-well plate were infected with serial tenfold diluted supernatant. CPE were observed and recorded at 96 h after infection. Virus titer was calculated by Reed–Muench method (Kint et al. 2015). As shown in Fig. 1E, all the seven isolated virus showed similar growth character: virus titers are detectable at 1 dpi, peak at 3 dpi, and then begin to decline. Among the seven isolates, the peak titer of isolate 12# reached 7.75 TCID50/mL, which is much higher than the other ones.
In conclusion, seven SARS-CoV-2 strains were isolated, sequenced and characterized in Vero cells, and a deletion mutation was identified after short passage in Vero cells. These results shall facilitate the understanding of the characteristics of SARS-CoV-2 in vitro.
HTML
-
This work was partially supported by the National Natural Science Foundation of China (31702248).
-
The authors declare that they have no conflict of interest.
-
The study was approved by the Ethics Committees of Prevention and Control of Infectious Disease of Zhejiang Province. All participants provided written informed consent. Written consents were obtained from all parents involved in the study.















 DownLoad:
DownLoad: