HTML
-
Lassa virus (LASV) belongs to the Mammarenavirus genus, Arenaviridae family, causing about 300, 000 cases of Lassa fever per year with the mortality rate in hospitals of 15% to 30% (Oldstone 2002; Houlihan and Behrens 2017). Mammarenaviruses are classified into the Old World (OW) arenaviruses and New World (NW) arenaviruses based mainly on virus genetics, serology, antigenic properties, and geographical relationships (Nunberg and York 2012). The OW LASV and Lujo virus (LUJV), as well as NW Junín virus (JUNV), Machupo virus (MACV), Guanarito virus (GTOV), and Sabiá virus (SBAV), are known to cause severe hemorrhagic fever and are listed as biosafety level 4 (BSL-4) agents (Buchmeier et al. 2007).
Arenaviruses are enveloped, negative-sense RNA viruses containing two genomic strands, S and L. L segment encodes the viral polymerase L and matrix protein (Z), while S segment encodes nucleoprotein (NP) and glycoprotein complex (GPC). During maturation, the precursor GPC is cleaved first by signal peptidase, followed with the cellular subtilisin kexin isozyme-1/site-1 protease, generating three noncovalently associated subunits: stable-signal peptide (SSP), GP1 and GP2 (Lenz et al. 2001). The unusually retained arenavirus SSP with 58 amino acids contains two transmembrane domains (TM) joined by an ectodomain loop containing 8 residues (Agnihothram et al. 2007). GP1 is responsible for the receptor binding. OW arenaviruses utilize α-dystroglycan (α-DG) as the primary receptor except for LUJV, which uses the transmembrane protein neuropilin 2 (NRP2) (Cao et al. 1998; Raaben et al. 2017). The pathogenic NW arenaviruses utilize human transferrin receptor 1 (hTfR1) as the receptor (Radoshitzky et al. 2007). The membrane fusion domain GP2 belongs to the class Ⅰ fusion protein (Igonet et al. 2011). Of note, the proximate external membrane region and TM of GP2, together with the ectodomain loop and TMs of SSP, form an SSP-GP2 interface, playing essential roles in regulating membrane fusion, and providing targets for distinct fusion inhibitors (York et al. 2004; York et al. 2005; Bolken et al. 2006; York and Nunberg 2006; Saunders et al. 2007; Larson et al. 2008; Lee et al. 2008; York et al. 2008; York and Nunberg 2009; Thomas et al. 2011; Messina et al. 2012; Burgeson et al. 2013; Ngo et al. 2015; Rathbun et al. 2015; Shankar et al. 2016; Wang et al. 2016; Wang et al. 2018).
To date, the high-resolution structure of the intact LASV GPC is not available, presumably due to the inherent metastability of the complex. LASV GPC is characterized as a class Ⅰ fusion protein, containing two heptad repeat (HR) regions, HR1 and HR2 in GP2. Once within the endosome, the GP1-GP2 complex undergoes conformational changes during endosome acidification. Eventually fusion activation leads to exposure of the fusion peptide, which is inserted into the cellular membrane. Finally collapse of the HR1 and HR2 regions forms a six-helix bundle (6HB), producing a fusion pore and the release of the ribonucleoprotein (Igonet et al. 2011). Formation of 6HB results in the fusion peptide and loop being in close proximity to the TM region. The adjacent MPER might contribute to the driving force to make the fusion event. The MPER of arenavirus, including the end of the external region of GP2 and the external region of SSP, might play critical roles in GPC-mediated function, such as sensing the low pH, triggering the membrane fusion, and targeted by the membrane fusion inhibitors. The eight amino acids in the external region of arenavirus SSP were investigated, and the conserved lysine at position 33 (K33) and asparagine at 37 (N37) were proven to be critical for GPCmediated cell fusion (York and Nunberg 2006; Saunders et al. 2007; York et al. 2008; York and Nunberg 2009). Here, we analyze the expression and functions of a panel of mutated LASV GP2 MPER with the aim of gaining a clearer understanding of the GP2 MPER functions. We find that mutations of four amino acid positions hamper LASV GPC-mediated membrane fusion.
-
HEK 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM; HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA). All point mutants were made from pCAGGS-GPC (LASV, strain Josiah, GenBank accession number HQ688673.1) by use of a Fast mutagenesis system (Transgene biotech), and mutations were confirmed by sequencing (Sangon, Shanghai) (Willard et al. 2018; Zhang et al. 2019). All transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
-
Equal amounts of cell lysates were subjected to SDSPAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore). Beta-tubulin was detected by using anti-β tubulin monoclonal antibody (AC021, Abclonal, ). LASV GPC and GP2 were detected by using rabbit anti-GP2 polyclonal antibody (1:1000 dilution) prepared by our group following reported methods (Eschli et al. 2006).
-
293T cells were transfected with pEGFP-N1 in combination with pCAGGS-LASV GPC or the empty pCAGGS. After 24 h, the cells were treated with acidified (pH 5.0) medium for 15 min. The cells were then placed in neutral medium, and syncytium formation was visualized 3 h later via fluorescent microscopy.
The quantification fusion assay was carried out as previously reported (Wang et al. 2018; Zhang et al. 2019). Briefly, cells transfected with both pCAGGS-LASV GPC and plasmids expressing T7 RNA polymerase were cocultured with targeted cells transfected with pT7EMCVLuc and pRL-CMV (plasmids used in this assay were kindly provided by Yoshiharu Matsuura, Osaka University, Osaka, Japan). After 12 h of incubation, the membrane fusion was triggered by treatment with acidified medium. Cell fusion activity was quantitatively determined after 24 h by measuring firefly luciferase activity expressed by pT7EMCVLuc and was standardized with Rluc activity expressed by pRL-CMV by using the Dual-Glo luciferase assay (Promega).
-
At 48 h post-transfection, cells were labeled with EZ-Link Sulfo-NHS-SS-Biotin working buffer (Thermo Scientific). After lysed with the Triton lysis buffer (25 mmol/L HEPES, 100 mmol/L NaCl, 1 mmol/L EDTA, 10% glycerol, and 1% Triton X-100), the biotin labeled surface protein was harvested by using NeutrAvidin Beads (Thermo Scientific). To assess the level of GPC, samples were subjected to Western blot (Eichler et al. 2006; CohenDvashi et al. 2016).
-
Student's t test was used to evaluate the statistical significance of differences. A value of P < 0.05 was considered statistically significant.
Construction of Mutants and Transfection
Western blotting
Membrane Fusion Assay
Detection of Cell Surface Protein
Statistical Analysis
-
Based on the recently released LASV GPC structure (PDB: 5VK2) (Li et al. 2016; Hastie et al. 2017), the end of HR2 is a glutamine at position 416. GP2 MPER located between the HR2 and transmembrane domain (TM), inclusive of residues 412–427 in the GP2 stalk (Fig. 1A, 1B). The fine structure of the stalk is not available (Fig. 1C). To examine the role of individual residues within the GP2 MPER regions, we preformed alanine scanning mutagenesis for each residue from T412 through P427.

Figure 1. Schematic diagrams of MPER of LASV GPC. A Schematic diagram of the LASV GPC open reading frame, with the cleavage sites of SPase and SKI-1/S1P indicated. The transmembrane domains in SSP and GP2 are shown in gray. The N- and C-terminal helices in GP2 are shown as lower diagonal and upper diagonal hatching, respectively. B Sequence alignment of LASV MPER aa 412–427 with other mammarenaviruses. C LASV GPC structure with the MPER highlighted in purple frame (EMD-3290) (Li et al. 2016).
To assess the role of MPER in precursor GPC (pGPC) expression and the maturation cleavage, 293T cells were transfected with mutated constructs. 24 h later, the corresponding cell lysates were subjected to Western blot assays by using anti-GP2 antibody. As shown in Fig. 2A, all mutant GPC proteins were expressed and processed properly into GP1 and GP2 subunits, although some proteins were expressed at reduced levels, such as T412A (Fig. 2B).
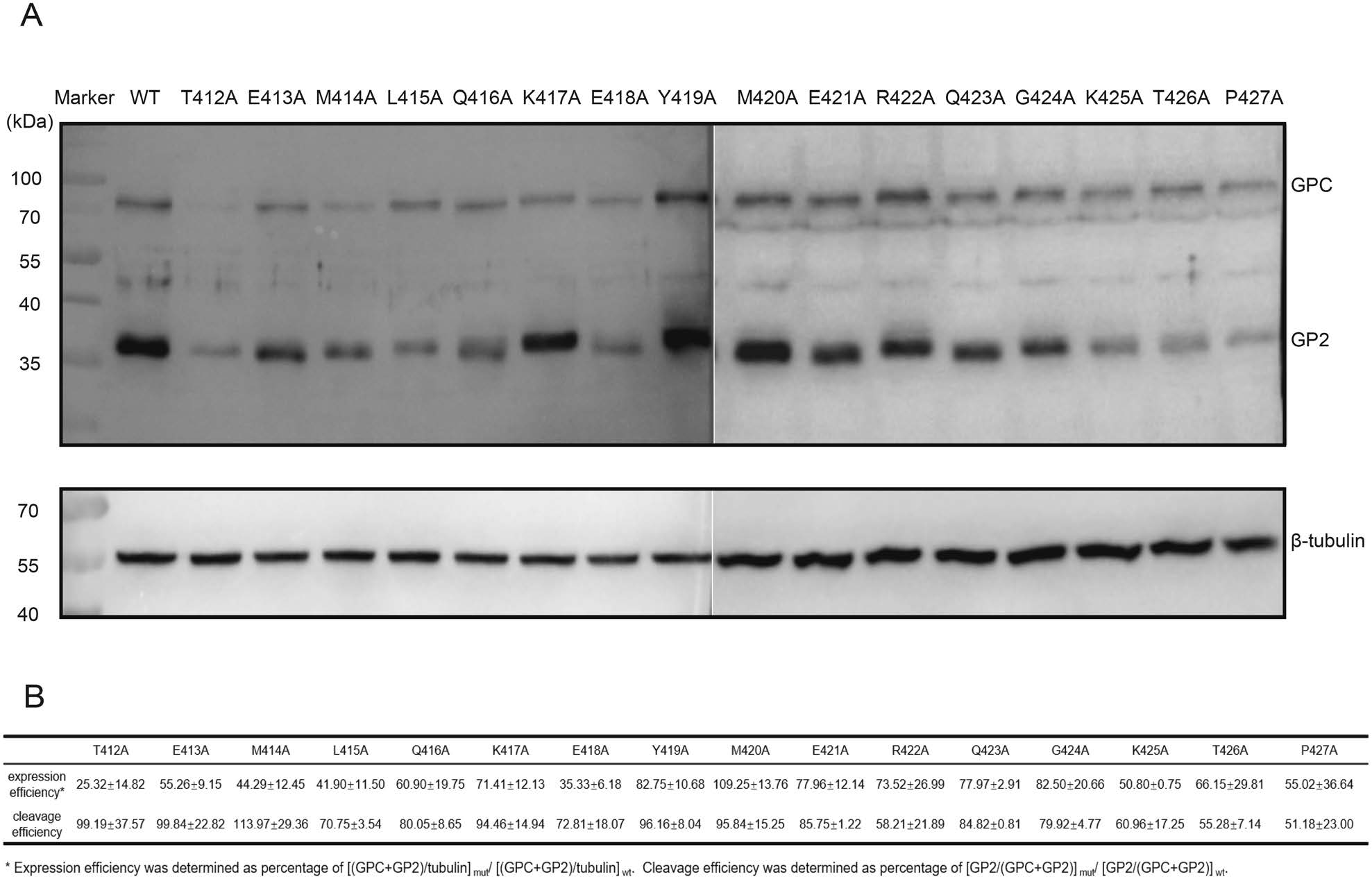
Figure 2. Proteolytic processing of MPER mutants. HEK 293T cells were transfected with WT or alanine mutants of LASV MPER. A The cell lysates were subjected to Western blotting by using anti-GP2 antibody. B Quantitative analysis of the expression and cleavage efficiencies based on the intensity of the bands.
-
To determine the importance of the residues within the MPER region to fusion activity, the fusion activity of the 16 alanine scanning mutants were examined (Wang et al. 2018; Zhang et al. 2019). Fusion activity was retained at most positions, with the exception that four mutations, M414A, L415A, K417A, and Y419A, resulted in nonfunctional GPCs that were fusogenically inactive, whereas the E418A, R422A, T426A and P427A mutants exhibited partially fusogenic phenotypes (Fig. 3A, 3B). Three of the mutations, L415A, K417A, and R422A were previously reported to impair GPC-mediated membrane fusion to < 30% level (York and Nunberg 2007; Willard et al. 2018), in which the fusion activities of L415A and K417A were similar to the data reported here, while the fusogenicity of R422A was quite different from our fusion assay. The discrepancy might be due to the differences of the experimental system.
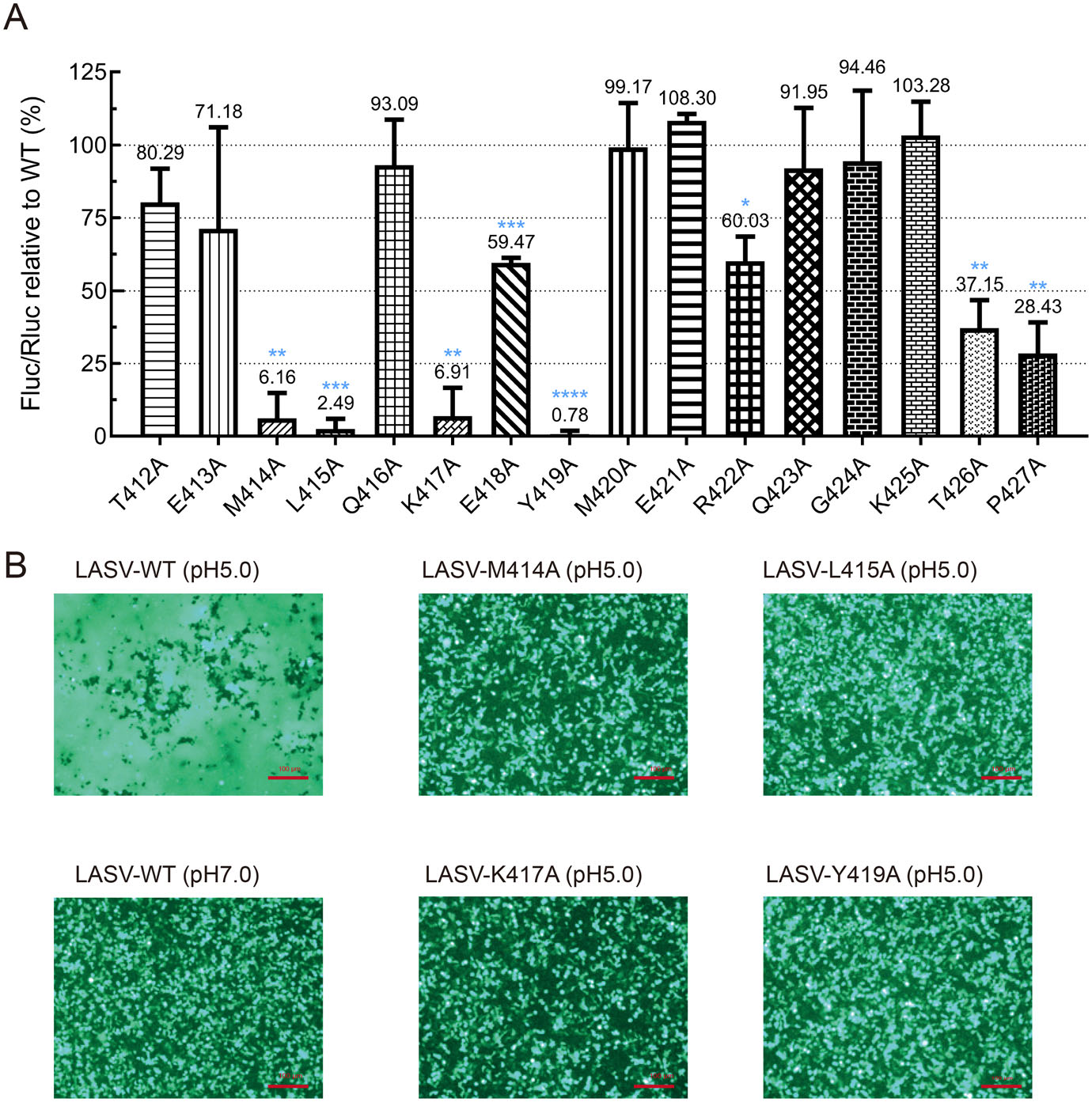
Figure 3. Fusion activities of MPER mutants. A Quantitate analysis of the fusion activities of each mutant in MPER. Error bars represent the SEM from at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. B The fusogenic phenomena mediated by the WT GPC was triggered by low pH, while it was blocked by transfected with M414A, L415A, K417A, or Y419A mutant.
The finding that K417 was crucial for the function was expected since the equivalent lysine residue (K403) in GTOV GP2 has been shown to form a salt bridge with the conserved glutamic acid and aspartic acid in the N-terminal of the adjacent HR1 of GTOV GP2, which contribute to the stabilization of the postfusion trimer conformation (Parsy et al. 2013). Although in the most recently published postfusion conformation of LASV GP2, this salt bridge was absent in the trimeric structure, the extended orientation of the HR2 of LASV GP2 allows them to interact closely with the HR1 of nearby neighbors (Shulman et al. 2019).
-
Intriguingly, it was also found that the residues with bulky side were key residues in MPER. Many factors may result in the loss of fusogenicity caused by the alanine mutant.Our fusion assay only monitors GPC refolding when present on the cell surface; to examine surface expression levels of the mutant GPC, we preformed cell surface biotinylation experiments. As shown in Fig. 4A, surface expression studies revealed that M414A, K417A and Y419A accumulated to similar levels as WT, whereas L415A mutant led to a reduction of mature GPC on the cell surface. As the surface transport is the basis for the subsequent fusogenicity, the loss of fusogenicity caused by L415A might be due to the loss of surface transport.
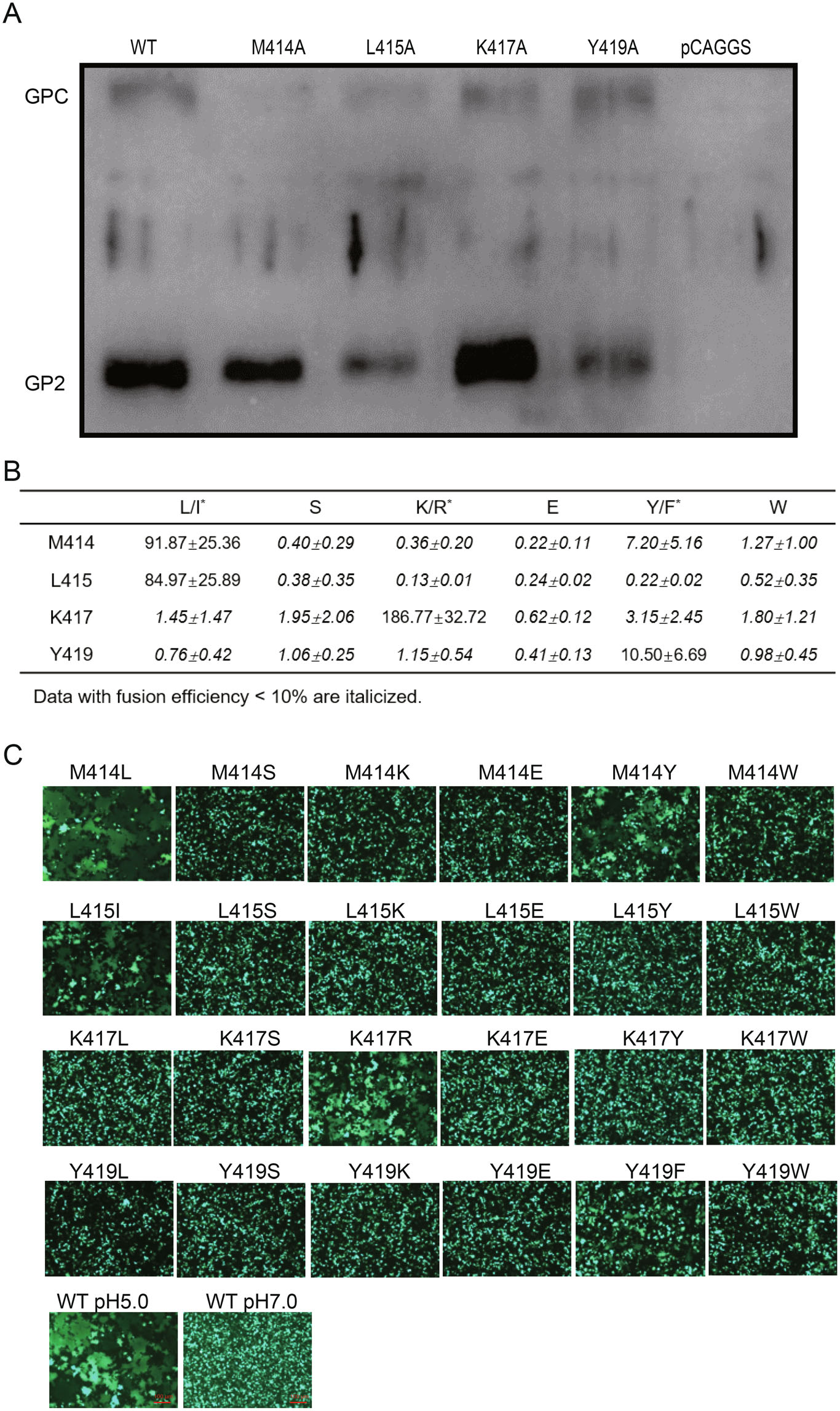
Figure 4. Characteristics of key residues for the role in fusogenicity. A Cell surface transport of WT and mutant GPCs. B Substitution of the four key residues with the similar amino acid partly retained the fusogenicity. Fusion efficiency was quantitatively determined by using the Dual-Glo luciferase assay. All data are displayed as a percentage of WT control ± SEM. C Images are representative fields from four to five independent experiments.
Bulky branched residues located in MPER have been shown to be essential to glycoprotein functions (Salamango and Johnson 2015; Lee et al. 2017; Pinto et al. 2019). The alanine substitution significantly truncates the side-chain length and decreases the hydrophobic effect, which might disrupt some critical contacts participated by the original bulky ones. To address it, we further replaced these four bulky residues into hydrophobic, polar, charged, and aromatic residues (Fig. 4B). As a result, substitution of M414 with either leucine or tyrosine could partly reconstitute the fusogenic function. It was noteworthy that LASV lineage Ⅱ (237-S-Nigeria- 2010H) presented leucine at position 414, so substitution of M414 of LASV lineage IV with the L of lineage Ⅱ could still lead to membrane fusion. Similarly, substitution of M414 with tyrosine could result in membrane fusion as well, while the M414S, M414K, M414E, and M414W failed; suggesting that position 414 was tolerable for amino acid with the similar size. It was notable that, for the other three investigated residues, only if replaced with the similar amino acid could partly restore the fusogenic effects. Only L415I was able to mediate fusion, indicating that side-chain length was indispensable at this position. Similarly, the positive charged residues were critical for position 417 because K417R was functional while others are not. It was also found that substitution Y419 with phenylalanine possessing the benzene ring retained the fusogenic effect (Fig. 4C).
Roles of GP2 MPER Mutations in GPC Expression and Cleavage
Roles of MPER Mutations on GPC-Mediated Membrane Fusion
Characteristics of Key Residues for the Role in Fusogenicity
-
Although the core structure of the LASV GP2 has been solved (Hastie et al. 2017; Hastie et al. 2019; Shulman et al. 2019), detailed structural information of LASV GP2 MPER in the GP2 stalk has been lacking. We scanned the MPER of LASV GPC to explore the role of these residues in glycoprotein expression, cleavage, and GPC-mediated fusion. MPER plays essential role in membrane fusion. The MPER of filovirus has been shown to interact with the fusion peptide to promote the membrane fusion (Lee et al. 2017). For flavivirus, which utilize the class Ⅱ fusion protein E as the glycoprotein, the MPER domain zip into the domain Ⅱ of E protein to trigger the membrane fusion (Chen et al. 2017). Although GPC belongs to the class Ⅰ fusion protein, the complex lacks a central three-helix fusion subunit core evident in other class Ⅰ fusion protein such as Ebola virus GP, HIV-1 Env, and influenza hemagglutinin (Hastie et al. 2017). We have tried to conduct combinatorial mutants in the N-terminal of GP2, such as D311K combined with K417E to explore the interaction of MPER with the N-terminal of GP2, however the additional D311K could hardly rescue the loss of fusion activity caused by K417E (data not shown). Whether the mutants tested in this study blocking the GPC mediated fusion by interrupting the interaction with the buried fusion peptide and HR1 of GP2 or by stabilizing the prefusion conformation of GPC by interacting with the ectodomain of SSP needs further elucidated.
The interface between the MPER/TM and the SSP of arenavirus GPC provides an Achilles' heel targeted by inhibitors. Residues in arenavirus GPC associated with resistance to or dependence on small-molecule fusion inhibitors are mostly located in this interface (Shankar et al. 2016; Wang et al. 2016; Zhang et al. 2019). The four key residues (M414, L415, K417, and Y419) reported here have not shown sensitivity or resistance to inhibitors (data not shown). They might serve as the scaffold to stabilize the structure of GPC rather than the direct viral target of inhibitors. Moreover, some mutant residues located in the MPER/TM would lead to the attenuated virions, and thus could be used for the design of vaccine. The live-attenuated Candid#1 vaccine is used in Argentina to protect against another high pathogenic arenavirus JUNV infection. Candid#1 attenuation is entirely dependent on F427I mutant in the N-terminal of the transmembrane domain of the JUNV GP2. Comparing the residues responsible for fusogenicity studied in this study to the corresponding sites in JUNV MPER, we found that some mutants caused different results in fusion assay, such as JUNV M406A (corresponding to LASV M414A) and K409A (LASV K417A) still led to > 50% fusogenicity, while JUNV E405A (LASV E413A) and R414A (LASV R422A) disrupt the fusogenicity (York et al. 2005; York et al. 2008), which suggested that specific atomic contacts in the two GPCs produce different results, and thus led to different virulence (York and Nunberg 2009).
Collectively, we scanned the LASV MPER which plays essential roles in membrane fusion, and some residues that possess the bulky branch or charge is indispensable for the GPC-mediated fusogenic activity.
-
We thank the Center for Instrumental Analysis and Metrology, Core Facility and Technical Support, and Center for Animal Experiment, Wuhan Institute of Virology, for providing technical assistance. This work was supported by the National Key Research and Development Program of China (2018YFA0507204), the National Natural Sciences Foundation of China (31670165), Wuhan National Biosafety Laboratory, Chinese Academy of Sciences Advanced Customer Cultivation Project (2019ACCP-MS03), the Open Research Fund Program of the State Key Laboratory of Virology of China (2018IOV001).
-
JC, GX, and WW conceived and designed the study. Experiments were performed by JC, GZ, MZ, and YL. Data were analyzed by JC and WW. Advice for the project was received from GX and WW. WW and JC wrote the manuscript. All authors read and approved the final manuscript.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.















 DownLoad:
DownLoad: