HTML
-
Severe acute respiratory syndrome-related coronavirus-2 (SARS-CoV-2), the causative agent of coronavirus disease-2019 (COVID-19), leads to one of the most serious worldwide pandemics in human history and is held responsible for over 100 million confirmed infections and more than 2 million fatalities by the end of January, 2021 (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). The lack of knowledge about this virus largely limits our capability to develop new therapeutic strategies, and thus it is urgent to improve the research on this contagious and lethal virus.
SARS-CoV-2 belongs to the Betacoronavirus genus of the Coronaviridae family, which wraps the largest known viral RNAs inside (Chen et al. 2020). The two-third of the viral genome at the 5′-proximal end encodes the viral replicase polyproteins 1a and 1b, the latter being translated by programmed -1 ribosomal frameshift. The one-third of the viral genome at the 3′-proximal end encodes ten proteins, including 4 well-known structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N), and a set of accessory proteins without clearly defined functions, which are expressed from the subgenomic viral RNAs generated through discontinuous transcription mechanism (Chen et al. 2020; Zhou et al. 2020).
Since the outbreak of COVID-19, a few drugs targeting various proteins coded by the viral genome were identified in the in vitro assays or cell culture with live virus; however, due to the lack of direct evidence of the inhibitory effect on the viral replication/transcription, the investigations on most of them were halted and most of them were proven inefficient or ineffective eventually in clinical trials (Hoffmann et al. 2020; Shrestha et al. 2020; Wang et al. 2020). For the assay on live SARS-CoV-2, the biosafety level 3 facility is the mandatory requirement. However, the biosafety level 3 laboratory is not widely available, and its shortage largely brings limitations to SARS-CoV-2 research and drug screening. In addition, work in the biosafety level 3 facility is also cost-expensive and laborious, and sometimes may have biosafety risks. Therefore, the generation of an easy-to-use and biosafe SARS-CoV-2 replication system is highly needed for current and future basic research and drug screening for eventual control of SARS-CoV-2 epidemic.
The biosafe viral replicon system is one of the best substitutes for contagious live virus. In such a system, it keeps the capability of genome or subgenome replication but it cannot produce infectious virus particles, thus avoiding possible biosafety risk of the living virus and making it possible to work in the biosafety 2 facilities. One of the successful examples is the establishment of hepatitis C virus (HCV) replicon, which contributes to the basic research and development of anti-HCV drugs and the final cure of the deadly disease caused by HCV (Lohmann et al. 1999; Lawitz et al. 2014).
Several strategies have been adopted to obtain a full-length infectious cDNA clone of coronaviruses. One of them is based on the ligation of viral cDNA fragments that contain the sticky ends generated by type ⅡS restriction enzymes (Hou et al. 2020; Xie et al. 2020a, 2020b), or in vivo recombination system to generate the full-length of viral cDNA in yeast cells (Thi Nhu Thao et al. 2020). Subsequently, the viral RNA is transcribed from the viral cDNA in vitro using T7 RNA polymerase and electroporated into mammalian cells to rescue the viral particles. The other strategy is to clone the viral full-length cDNA into one plasmid and the viral cDNA expression is driven directly by mammalian polymerase Ⅱ promoter such as human cytomegalovirus (CMV) promoter in mammalian cells (Almazan et al. 2006). Full-length infectious cDNA clones have been established for SARS-CoV-2 by several research groups, which lead to the production of recombinant infectious SARS-CoV-2 (Hou et al. 2020; Thi Nhu Thao et al. 2020; Xie et al. 2020a). However, a biosafe and robust SARS-CoV-2 replicon is still lacking.
Previously, we have generated a biosafe and reporter gene-containing SARS-CoV replicon by deleting all the structural and accessory genes of SARS-CoV, excepting that for N protein (Almazan et al. 2006; Pan et al. 2008). In this study, we generated a SARS-CoV-2 cDNA clone in the vector of bacterial artificial chromosome (BAC) with deletion of the whole S gene fragment and insertion of reporter gene at the S locus. Transfection of this cDNA clone led to robust replication of SARS-CoV-2 replicon RNA but did not produce infectious virus particles due to the lack of S gene. We demonstrated that such a replicon system could be used for studies of SARS-CoV-2 replication/transcription, including the functions of the viral RNA-dependent RNA polymerase (RdRP), exonuclease, and cap N7 methyltransferases. We also showed that the replicon could be used for testing the inhibitors and antiviral drugs that target SARS-CoV-2 replication/transcription apparatus, indicating that it can serve as a biosafe and efficient tool for both basic research and development of anti-SARS-CoV-2 drugs.
-
HEK293T, Huh-7, A549, BHK-21, DLD-1, SY-SH5H, and Vero E6 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin (Thermo Fisher Scientific, Waltham, MA, USA). Sixteen to twenty-four hours before transfection, 1 × 106 HEK293T, 2 × 106 Huh-7, 2 × 106 A549, 1.5 × 106 BHK-21, 1 × 106 DLD-1, 1.5 × 106 SY-SH5H, and 1 × 106 Vero E6 cells are plated in 6-cm plate. Two hours before transfection, the medium is replaced with DMEM without serum and antibiotics. For each 6-cm plate, 3 μg replicon plasmid and 6 μL Hieff TransTM Liposomal Transfection Reagent (Yeasen Biotech, Cat#40802ES03, Shanghai, China) were used for transfection. Six h post-transfection, the medium containing transfection mixture is replaced with DMEM supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin. Forty-eight hours post-transfection, the cells were subjected to various assays.
-
To utilize the elements of transcription system of mammalian cells to drive the synthesis of RNA of SARS-CoV-2, we installed CMV promoter upstream of and ribozyme, poly-A, and BGH elements downstream of viral cDNA sequence. To eliminate the potential infectivity of viral RNA, we removed the coding sequence of S gene. The replicon sequence containing S gene-deleted viral cDNA and essential elements for transcription was divided into 7 fragments which were synthesized by GENEWIZ (Suzhou, China). Fragment 1, 2, 3, 5, 6, and 7 were cloned into pUC57 vector and fragment 4 was in pCC1BAC vector. All the fragments were amplified by PCR (Supplementary Table S1) and digested with BsmBI except for fragment 1 with both BsmBI and KasI and fragment 7 with both BsmBI and HindⅡI. Fragment 7 was the first one inserted in pBAC-MCS vector with designed restriction sites and then the other fragments could be cloned in the vector in any order. The ligation products were transformed in chemically competent DH10B cells and the positive clones were screened by colony PCR. The clones were amplified and the plasmids were extracted with NucleoBond BAC 100 (Takara Bio, Dalian, China). All clones were validated by Sanger sequencing (TSINGKE biotech, Guangzhou, China).
-
The assays were performed following the manufacturer's instructions (Promega Corporation, Fitchburg, WI, USA). In brief, the cells in 12-well plate transfected with 500 ng pBAC-nCoV-Replicon-Luciferase plasmid and 50 ng RL-TK plasmid expressing renilla luciferase were lysed in 200 μL 1× Passive Lysis Buffer (PLB). 20 μL of each lysate was transferred into 96-well white plate and then mixed with 20 μL Luciferase Assay Reagent Ⅱ and subsequent with 20 μL Stop & Glo solution. The luminescence values of two steps of reactions were recorded using luminescence detector in Synergy H1 Hybrid Multi-Mode Reader (BioTek, Winooski, VT, USA).
-
Microscopy was performed as previously described (Pan et al. 2011). In brief, the cells were plated in 12-well plate with a coated coverslip on the bottom. 48 h post-transfection, the cells were fixed with 4% paraformaldehyde solution and subsequently permeabilized with PBS containing 1% TRITON-X-100. Before the coverslip is set on the slide with Immu-Mount (Thermofisher Scientific), the cells were stained with DAPI (1 μg/mL). The slides were observed and imaged using Nikon DS-Qi2. The green cells in four independent representative areas were counted.
-
The viral genomic cDNA sequence is separated by unique restriction sites, KasI, NheI, PacI, ClaI, MluI, AxyI, SacⅡ and AscI. The fragment containing the mutant sites and flanked by respective restriction sites was amplified following the common strategy of over-lap PCR. Briefly, the complementary forward and reverse primers containing the mutant sites are paired with the primers downstream or upstream the restriction nucleotides (Supplementary Table S2). Two PCR products amplified by the two primers were then used as the template for the second round of PCR. The nest-PCR strategy was employed to improve the production of PCR products. The replicon plasmid and PCR product were digested with the respective restriction enzymes, gel-purified, and ligated at 4 ℃ overnight. The chemically competent DH10B cells are transformed with ligation samples and positive clones are screened with the strategy of colony PCR. The sequence is verified with Sanger sequencing to ensure no undesirable mutations.
-
Total RNAs were extracted from cells using Trizol reagent (Thermo Fisher Scientific, Cat#15596026) by following the manufacturer's instructions. The quality of each RNA sample was validated using agarose gel electrophoresis and NanoDrop One (Thermo Fisher Scientific). 2 μg of each RNA sample were reversely transcribed using a cDNA synthesis kit (Thermo Fisher Scientific, Cat#K 1622). The cDNA was subjected to real-time quantitative PCR (qRT-PCR) assay with 10 μL reaction mixture containing 5 μL TB GreenⓇ Premix Ex TaqTM (Takara Bio, Cat#RR420A). The assay was performed in ABI Detection System, ABI 7500, using the following programs: 30 s at 95 ℃, followed by 40 cycles of 5 s at 95 ℃ and 10 s at 60 ℃. The 2-∆∆CT method was used to calculate the relative gene expression of each sample.
-
Total RNA was extracted from HEK293T cells transfected with replicon or Vero E6 cells infected with virus. Low-concentration RNA libraries for each sample were prepared with Trio RNA-Seq kit (NuGEN Technologies, Redwood City, CA, USA). The libraries were then subjected to 150 bp pair-end sequencing with an Illumina Miseq platform. To estimate the composition of subgenomic RNAs within the sample, we counted the reads containing unique signature leader (11 base pair)—body (10 base pair) junction sequence (Supplementary Table S3) associated with each subgenomic RNA species. As a comparison, the compositions of subgenomic RNAs were also calculated from total RNA sequencing results of (1) cells infected with SARS-CoV-2 viruses and (2) respiratory tract samples from 10 infected patients.
-
Student's t-test was utilized to calculate the significance of the differences between two groups. Results were considered significant when P value was less than 0.05.
Cell Cultures and Transfection
Cloning of SARS-CoV-2 Replicon
Luciferase Assay
Microscopy
Mutagenesis
RNA Extraction and Real-Time Quantitative PCR
Quantification of Subgenomic RNAs by Deep Sequencing
Statistics
-
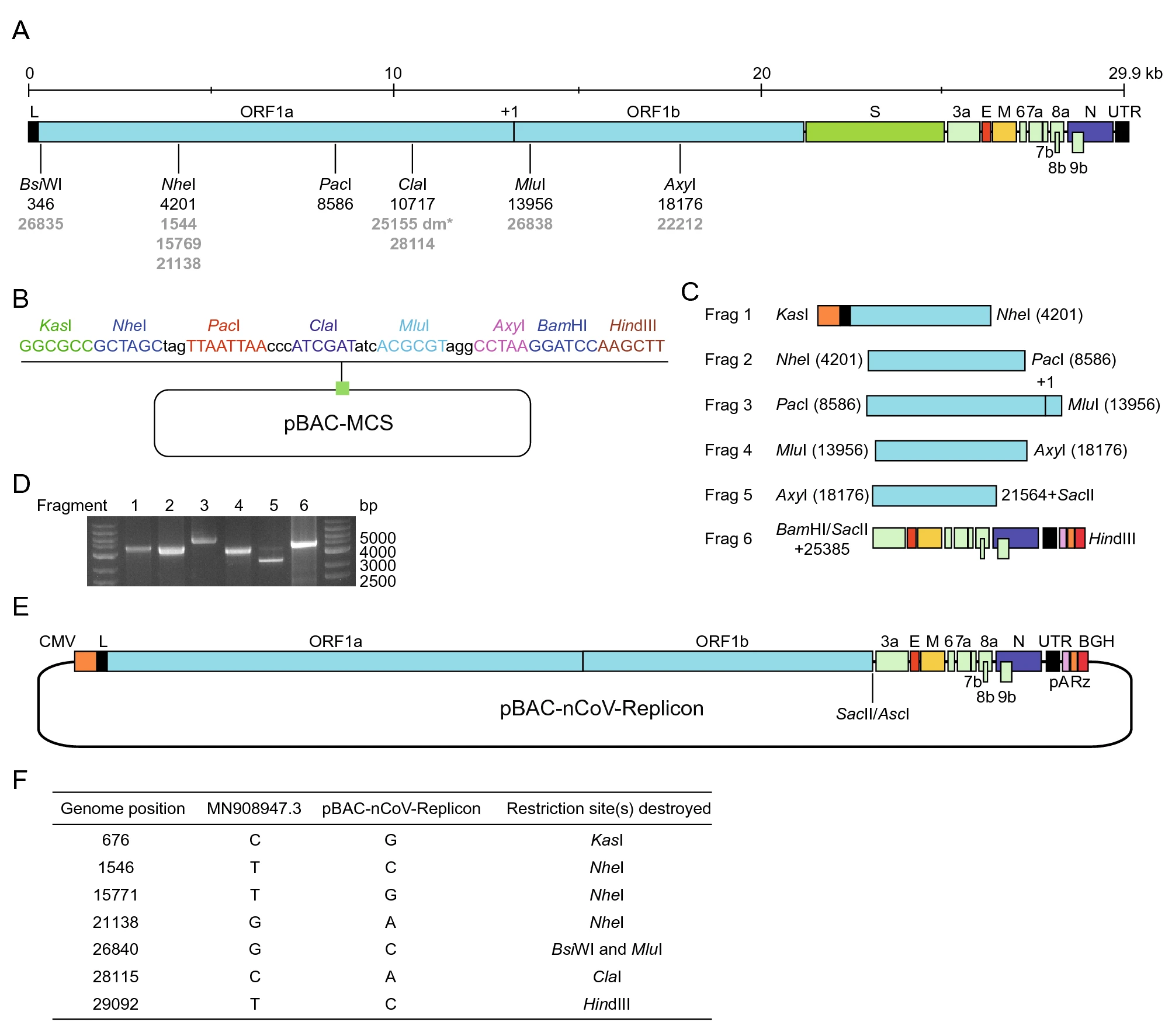
The viral cDNA sequence used for construction of the viral replicon is based on the strain of SARS-CoV-2 Wuhan-Hu-1 (MN908947.3), which is among the earliest ones characterized after the outbreak of COVID-19 (Wu et al. 2020). We adopted the in vitro ligation strategy to obtain this viral cDNA (Yount et al. 2000). For the construction of biosafe replicon system, we replaced the coding region of spike (S) gene, which is indispensable for the generation of infectious viral particles (Hierholzer et al. 1972; Li et al. 2003; Pan et al. 2008), with two restriction sites, SacⅡ and AscI, and retained its upstream transcription regulatory sequence (TRS). To use the transcription system for the synthesis of viral RNAs in the mammalian cells, we employed CMV promoter to drive the expression of viral cDNA and employed 25-nt synthetic adenine bases (pA), hepatitis delta virus-derived ribozyme (Rz) and the bovine growth hormone (BGH) termination and polyadenylation sequences to terminate the transcription. These elements were designed and utilized to produce authentic 5′ and 3′ ends of viral genome of SARS-CoV (Almazan et al. 2006; Pan et al. 2008). We separated the SARS-CoV-2 cDNA and functional elements into seven fragments for cDNA synthesis (Supplementary Fig. S1A, S1B). Each fragment is flanked at both ends with BsmBI, the type ⅡS restriction enzyme that generates unique sticky ends, except for fragment 1 with KasI upstream of CMV promoter and fragment 7 with HindⅡI downstream of BGH termination signal sequence. Each fragment was digested with respective restriction enzymes, gel-purified, and then ligated to assemble the cDNA sequence of replicon in vitro (Supplementary Fig. S1B). For the convenience of cloning manipulations, we introduced seven silent nucleotide changes in the viral cDNA as molecular markers, which destroyed the unfavorable restriction sites of KasI (676 nt), NheI (1546 nt, 15,771 nt and 21,138 nt), BsiWI and MluI (26,840 nt), ClaI (28,115 nt), and HindⅡI (29,092 nt) (Supplementary Fig. S1C, Fig. 1F).

Figure 1. Construction of a SARS-CoV-2 replicon. A Schematic of the genome of SARS-CoV-2 and restriction sites selected for cloning. B The cloning vector for replicon with designed restriction sites. C Schematic of the fragments for cloning with indicated restriction sites, elements for transcription, and positions in the viral genome. D The fragments for cloning were examined in agarose gel electrophoresis. E The organization of the SARS-CoV-2 replicon. Note that SacⅡ and AscI restriction sites were inserted downstream the TRS of S gene which is deleted to eliminate the ability of replicon to generating live virus. F The mutations destroying the unfavourable restriction sites were used as cloning markers.
-
BAC was employed as the backbone for cloning the cDNA of coronaviruses previously and it maintains the stability of cDNA clone for more than 200 generations (Almazan et al. 2006; Pan et al. 2008). It can be easily manipulated and amplified in bacteria, transfected and expressed in mammalian cells, and therefore, the replicons based on BAC have advantages over other approaches. In our laboratory practice, the construction of cDNA sequence with the size of up to 6 kb in BAC is performed routinely, and thus we divided the replicon sequence into fragments with the size of 4–5 kb (Fig. 1A). To insert SARS-CoV-2 replicon DNA sequences into the BAC vector, we generated a BAC vector containing desired restriction sites (pBAC-MCS) (Fig. 1B). The fragments of KasI-NheI, NheI-PacI, PacI-MluI, MluI-AxyI, AxyI-SacⅡ, and BamHI-HindⅡI were amplified from the corresponding cDNA sequence, digested with respective restriction enzymes, and gel purified (Fig. 1C, 1D). The fragment of BamHI-HindⅡI was inserted into pBAC-MCS at the first step and then all the other fragments could be inserted in any order. After all the fragments were assembled in pBAC-MCS (Fig. 1E), we sequenced the final construct to ensure no undesired mutations (Supplementary Table S4).
-
Production of a set of subgenomic RNAs is the remarkable feature of the coronavirus life cycle and the convincing indicator of viral replication (Hussain et al. 2005). Although it drives the generation of replicon genomic RNAs, the CMV promoter cannot promote the production of subgenomic RNAs (sgRNAs) because the coronaviruses synthesize their subgenomic RNAs via a discontinuous mechanism by their own replication/transcription complex. The sgRNAs comprise the same 5′-proximal leader sequence of 75-nt and 3′-body sequences of different lengths depending on the respective transcription regulatory sequences (TRS; its core sequence is ACGAAC for SARS-CoV-2) in the viral genome. Thus, the presence of sgRNAs in cells is indicative of coronavirus replication.
To demonstrate the replication and generation of sgRNAs of the replicon, we transfected HEK293T cells with replicon plasmid using a liposomal transfection reagent. Two days post-transfection, the cells were collected for RNA extraction and subsequent reverse transcription. We designed a set of primers for amplification of sgRNAs, including one universal primer in the leader sequence and the other primers in the various ORFs at 3′-proximal region of the viral genome (Supplementary Table S5). When setting the extension time of PCR program to 1 min, PCR products were detected with different lengths related to various species of subgenomic RNAs (Fig. 2A). However, when decreasing the extension time to 5 s, we could mainly detect the PCR products from the template composed of a direct fusion of leader sequence and coding regions of ORFs where the primer sequences are located (Fig. 2B), indicating that the primers could be reliably used to quantify the respective subgenomic RNAs.
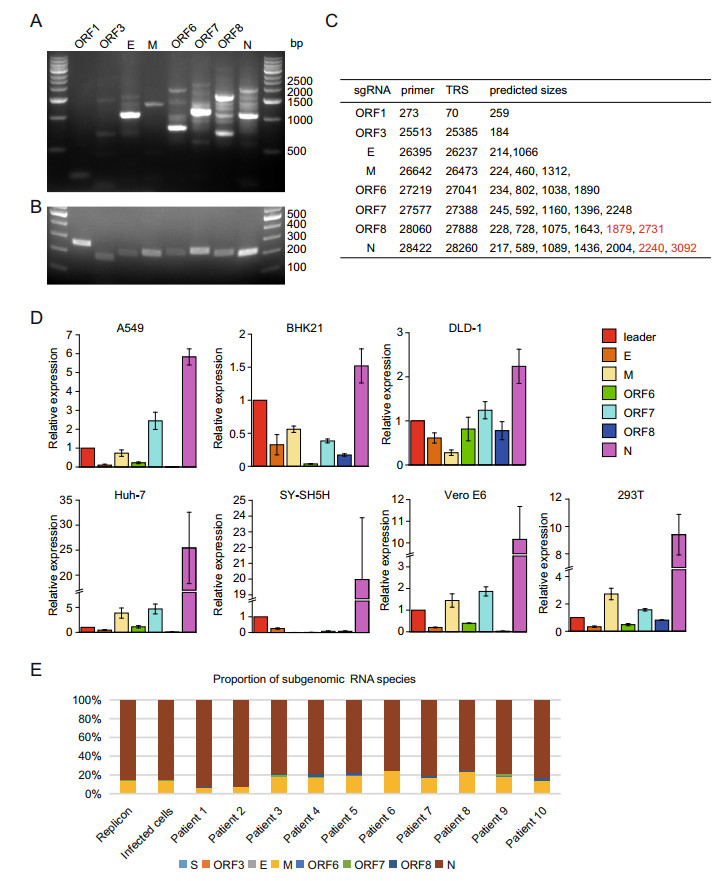
Figure 2. The replication and transcription of the SARS-CoV-2 replicon. The subgenomic RNAs from 293T cells transfected with replicon plasmid were examined using reverse-transcription PCR. The extension time of 1 min (A) and 5 s (B) were performed and the PCR products were detected with DNA agarose electrophoresis. The lengths of PCR fragments which could be amplified theoretically by different primers were depicted in the Table (C). Note that the fragments undetected by PCR were highlighted with red color. D The total RNAs from A549, BHK-21, DLD-1, HEK293T, Huh-7, SY-SH5H, and Vero E6 transfected with the replicon plasmid were reversely transcribed and the cDNA samples were subjected to qRT-PCR. The relative expressions of subgenomic RNAs of each cell line were presented. E Comparisons of the subgenomic RNA composition among Vero E6 expressing replicon, SARS-CoV-2-infected Vero E6 cells, and respiratory tract samples from ten SARS-CoV-2 infected patients. The relative proportion of each subgenomic RNA species was calculated by counting signature leader-body junction sequence for each subgenomic RNA species based on total RNA sequencing of the various samples, respectively.
Paired with the universal primer located in the leader sequence, the primers located in 3′-proximal ORFs could distinguish various sgRNAs and generated DNA products of different lengths in PCR reaction. The predicted sizes of PCR products were depicted in Fig. 2C. As shown in Fig. 2A, all the expected PCR fragments could be readily detected in the agarose gel after DNA electrophoresis, indicating that the transcription and replication of the SARS-CoV-2 replicon are fully functional in the mammalian cells. Possibly due to low amount or low efficacy in amplification of PCR, four fragments with the larger size (marked in red in Fig. 2C), i.e. 1879 bp and 2731 bp of ORF8 and 2240 bp and 3092 bp of N, could not be unambiguously identified in the agarose gel. This is not unexpected, for it happened in our previous work for the detection of sgRNAs of coronaviruses from the samples infected by live SARS-CoV by using the same strategies (Hussain et al. 2005).
Since various tissues and organs were reported to be infected with SARS-CoV-2 (Du et al. 2020; Paniz-Mondolfi et al. 2020) and the pathological outcomes varied markedly, we examined the transcription/replication of our replicon in cells from various tissues, A549 for lung, DLD-1 for colon, Huh-7 for liver, SY-SH5H for neuron, and 293T for kidney. The subgenomic RNAs were detected in all of these cells after the transfection with replicon plasmid (Fig. 2D). However, the proportions of various subgenomic RNAs in different cell lines varied markedly, suggesting that the viral transcription and replication could be affected by the host factors and this effect could contribute to the varied pathological outcomes in different tissues.
Next, we compared the replication and sgRNA production of the replicon with that of a live wild-type virus. The RNA samples were prepared from the cells transfected with replicon or infected with live viruses, and the respiratory tract samples from ten SARS-CoV-2-infected patients and subjected to 150-bp pair-end sequencing with an Illumina MiSeq platform. The results showed that the replicon and wildtype virus have similar types and proportions of the subgenomic RNA when replicating in Vero E6 cells (Fig. 2E), indicating that the replicon has a similar replication/transcription profile in host cells. However, the proportions of sgRNAs of the patient samples varied markedly, which may reflect the difference of disease progression stages or varied qualities of the patient samples (Fig. 2E). Together, our data demonstrated that the SARS-CoV-2 replicon could replicate robustly in cells and its replication/transcription profile was comparable to that of a natural live virus.
-
To further confirm that the generation of sgRNAs was the viral replicase activity-dependent, we introduced mutations at the key residues of replicase proteins to diminish the activities of the RdRP (nsp12 S759A/D760A/D761A, SDD), exonuclease (nsp14 D90A/E92A), or cap N7 methyltransferase (nsp14 D331A) in the replicon (Chen et al. 2009; Gao et al. 2020) (Fig. 3A). All three mutations drastically decreased the copy number of sgRNAs, indicating the impaired capability of replication/transcription of the replicon due to the introduced mutations (Fig. 3B). The production of sgRNAs of M and ORF7 are decreased as many as 100 times by SDD mutation. SDD mutation in RdRP leads to more impaired capability of replication/transcription compared with D90A/E92A or D331A mutation, indicating that exonuclease or cap N7 methyltransferase plays a less essential role in viral replication/transcription (Chen et al. 2009). In contrast, the generation of replicon genomic RNAs was less affected compared with sgRNAs (Fig. 3B with "leader"), and this can be explained by the fact that the synthesis of replicon genomic RNAs is driven by both the CMV promoter-mediated transcription from the replicon plasmid and the viral replicase-mediated transcription from the viral RNA template in the host cells. Therefore, when the replicon RNA replication and transcription were blocked, the replicon genome RNA, but not the subgenomic RNAs, could still be produced via host transcription mechanism. Collectively, these results showed that the synthesis of sgRNAs is mainly dependent on the viral transcription/replication mechanism, where RdRP, exonuclease, and cap N7 methyltransferase play important roles and the amounts of sgRNAs instead of replicon genomic RNAs manifest the replicon activity of replication/transcription. The mutational analysis also showed that such a replicon system can be adopted for basic research related to SARS-CoV-2 genome replication and transcription.
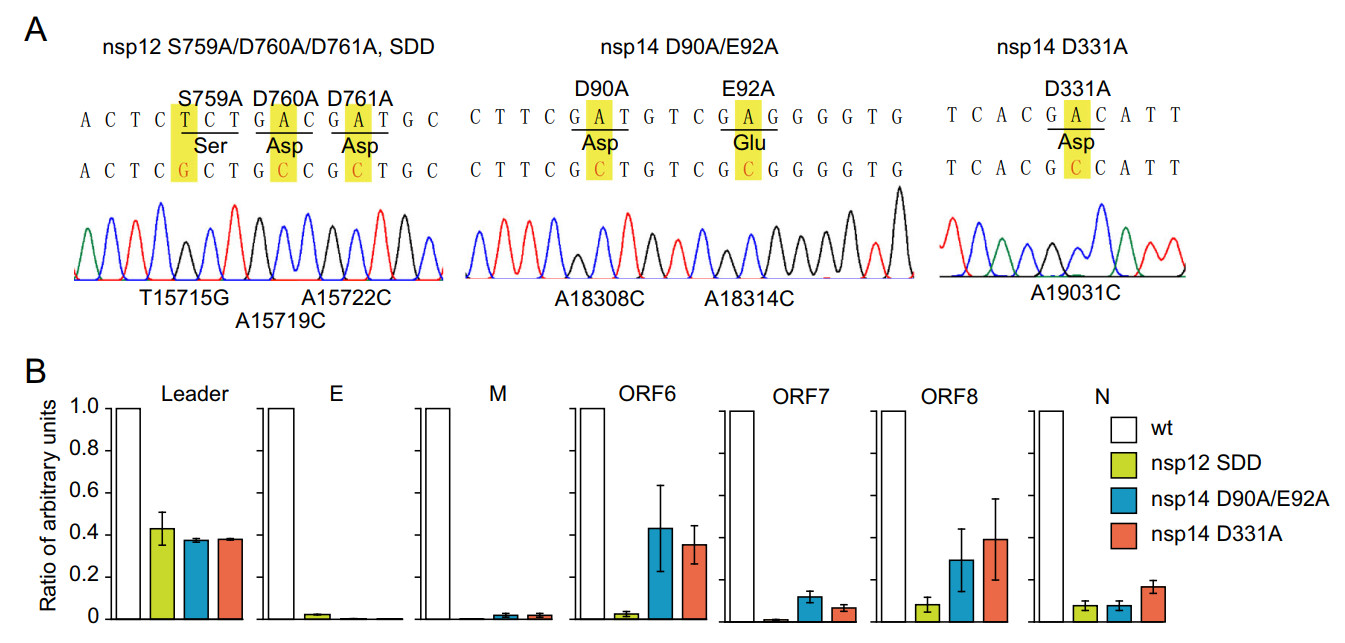
Figure 3. The mutations of viral non-structural proteins decrease the synthesis of subgenomic RNAs of the replicon. A The replicon with mutant sites in RdRP (S759A/D760A/D761A, SDD), exonuclease (D90A/E92A, DE), and cap N7 methyltransferase (D331A) were confirmed by Sanger sequencing. B HEK293T cells were transfected with wild-type or mutant replicons with RdRP (S759A/D760A/D761A, SDD), exonuclease (D90A/E92A, DE), or cap N7 methyltransferase (D331A). The relative quantity of subgenomic RNAs were analyzed by qRT-PCR. Note that mutations of RdRP (S759A/D760A/D761A, SDD), exonuclease (D90A/E92A, DE), or cap N7 methyltransferase (D331A) in replicon drastically decreased the synthesis of subgenomic RNAs.
-
Viral replicons with reporter gene were widely used as the model for large-scale screening for antiviral drugs (Hannemann 2020). To insert reporter gene into replicon, we designed SacⅡ and AscI restriction sites downstream of the TRS sequence of the S gene. We inserted luciferase, mNeonGreen, or puromycin gene between those two sites respectively (Fig. 4A) and the expressions of those genes are driven by transcription regulatory sequence of the S gene. After transfection using a liposomal reagent, the expressions of reporter genes were examined by the fluorescence measurement for mNeonGreen and the enzymatic activity assay for luciferase (Fig. 4B–4D). Consistent with the reduced production of sgRNAs (Fig. 3B), the mutations (SDD, D90A/E92A and D331A) also drastically diminished the expression of luciferase (Fig. 4B), and SDD mutation induced more decrease in the reporter activity of luciferase compared with that of D90A/E92A or D331A mutation.
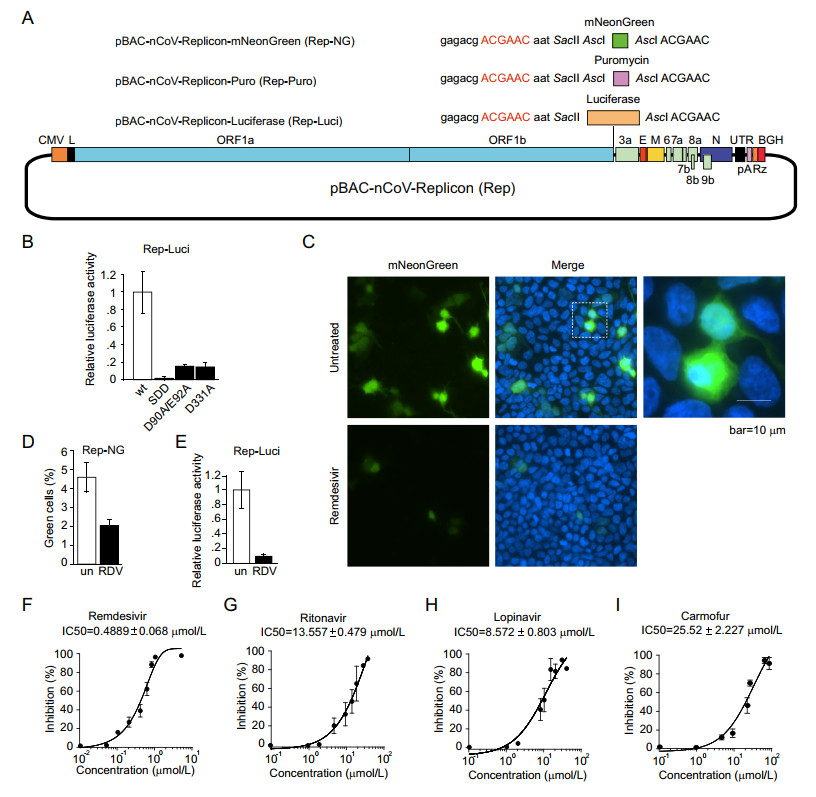
Figure 4. SARS-CoV-2 replicon functions as an efficient drug screening system. A The reporter gene, mNeonGreen or luciferase, were inserted between SacⅡ and AscI to obtain the constructs of pBAC-nCoV-Replicon-mNeonGreen (Rep-NG) and pBAC-nCoV-Replicon-Luciferase (Rep-Luci). The selection gene, puromycin, was inserted in AscI obtain the constructs of pBAC-nCoV-Replicon-puromycin (Rep-Puro). B HEK293T cells were transfected with wild-type or indicated mutant pBAC-nCoV-Replicon-Luci (Rep-Luci) and pRL-TK. 48 h post-transfection, the cells were subjected to the Dual-LuciferaseⓇ Reporter (DLRTM) Assay. HEK293T cells transfected with Rep-NG (C) and Rep-Luci (E) were left untreated or treated with Remdesivir (RDV) at 1 μmol/L. The cells transfected with Rep-NG were fixed and observed under microscope (C) and the green cells were counted in four independent areas (D). The cells transfected with Rep-Luci were subjected to Dual-LuciferaseⓇ Reporter (DLRTM) Assay (E). HEK293T cells transfected with Rep-Luci were treated with various drugs, Remdesivir (F), Ritonavir (G), Lopinavir (H) and Carmofur (I) with known inhibitory effect on the replication of SARS-CoV-2 at 8–9 different concentrations. The values of IC50 of each inhibitor were analyzed with Graphpad Prism.
To test whether our replicon with reporter gene is suitable for large-scale anti-viral drug screening, we used various known anti-SARS-CoV-2 drugs to treat the cells transfected with the replicon carrying a reporter gene. After the transfection of HEK293T cells with pBAC-nCoV-Replicon-mNeonGreen (Rep-NG), the green fluorescence was visible in around 5% cells. The percentage of visible green cells is decreased to less than 2% after the treatment of Remdesivir (Fig. 4C, 4D), and the intensities of green fluorescence in most of the cells were decreased drastically (Fig. 4C). We did the same treatment of Remdesivir on the cells transfected with pBAC-nCoV-Replicon-Luciferase (Rep-Luci) and the luciferase activity is decreased by more than 90% in comparison with untreated replicon-containing cells (Fig. 4E), which is consistent with the results from Rep-NG (Fig. 4C, 4D).
Next, we further investigated the inhibitory effect of various drugs on viral replication/transcription using our replicon system (Rep-Luci). We tested four known drugs that inhibit the replication/transcription of coronaviruses, including Remdesivir (an inhibitor for RdRP of coronaviruses and several other RNA viruses; Wang et al. 2020) (Fig. 4F), Ritonavir and Lopinavir (HIV protease inhibitors; Ho et al. 1995; Carrillo et al. 1998) (Fig. 4G, 4H), and Carmofur (an inhibitor for the main protease of SARS-CoV-2; Jin et al. 2020) (Fig. 4I). We tested various dosages of the four drugs on the Rep-Luci and observed the inhibitory effect at the dosages which do not cause cytotoxicity. Remdesivir showed a strong inhibitory effect (IC50 of 0.4889 μmol/L) on replicon replication/transcription, which was in agreement with other studies with the live virus (Wang et al. 2020). Lopinavir (IC50 of 8.572 μmol/L), Ritonavir (IC50 of 13.557 μmol/L), and Carmofur (IC50 of 25.52 μmol/L) also showed the inhibitory effect to some extent in the SARS-CoV-2 Rep-Luci reporter system (Fig. 4F–4I), consistent with the studies with live SARS-CoV-2 (Jin et al. 2020; Kang et al. 2020; Pizzorno et al. 2020). Collectively, the biosafe replicon is a reliable and efficient model for the detection of antiviral activity of drugs and thus an ideal platform for large-scale drug screening.
Design of a SARS-CoV-2 Replicon
Construction of a One-Plasmid SARS-CoV-2 Replicon in BAC Vector
SARS-CoV-2 Subgenomic Replicon Replicates Efficiently with Similar Replication/Transcription Profile to that Wild-Type Virus in Mammalian Cells
Mutational Analysis Demonstrated the Important Roles of RdRP, Exonuclease, or Cap N7 Methyltransferase Activity in Viral Replication/Transcription
Validation of SARS-CoV-2 Replicon as an Efficient Drug Screening Platform
-
Reverse genetics system for viral replication/transcription plays an important role in viral research and antiviral drug screening. The biosafe replicon model developed from reverse genetics system can reduce the usage of live viruses and make the research safer, especially for the viruses which require the laboratory of biosafety level 3. One of the successful examples is HCV replicon system, which was widely employed in HCV research and antiviral drug development. This HCV replicon contributes greatly to the generation of anti-HCV drugs such as ledipasvir and sofosbuvir, and the final cure of this disease caused by HCV (Lohmann et al. 1999; Lawitz et al. 2014). SARS-CoV-2 is highly contagious and the research on this virus is strictly limited in the facility of biosafety level 3. Thus, it is urgent to generate a biosafe replicon which on one hand reduces the concerns over the biosafety of the research work on this virus, and on the other hand, expands the accessibility to SARS-CoV-2 research for non-virologists. In this study, we filled in the gap and met the need for a biosafe SARS-CoV-2 replication system.
The coronaviruses possess the largest viral genomic RNAs and this makes it a big challenge to clone the full-length of viral cDNA into a single plasmid. To bypass this difficulty, the researchers designed the strategy of assembly of viral full-length cDNA from several viral fragments using in vitro ligation or in vivo yeast recombination system (Hou et al. 2020; Thi Nhu Thao et al. 2020; Xie et al. 2020a, 2020b). In this strategy, the viral RNA is transcribed from viral cDNA using a T7 transcription system and then delivered into mammalian cells using electroporation. This strategy is used mainly for generating recombinant infectious virus and it is difficult to construct a biosafe replicon by using this strategy because the efficiency of replicon RNA transfection and rescue in the cells is relatively low and unstable. In addition, it is not convenient for many laboratories without expertise in handling the large DNA fragments for in vitro ligation or recombination, in vitro production of viral RNAs, and electroporation with RNA samples. Therefore, in the current study, we aimed to generate a bio-safe and easy-to-use SARS-CoV-2 replicon, which is carried in one plasmid and can initiate the replication and transcription after transfection into the cells.
To construct this replicon, we employed the BAC vector which was widely used for cloning large DNA fragments and the CMV promoter which efficiently drives the transcription of large RNAs. To assure the biosafety of the replicon, we left out the S gene in the viral cDNA and abolished the capability of the generation of live viral particles. To validate the activity of the replication/transcription of our replicon, we examined the expressions of various sgRNAs of the replicon and proved the profile of the expression of the subgenomic RNAs is similar to that of live virus. Our data showed that the replicon could be a perfect substitute for live virus without causing the biosafety concerns.
For convenient manipulation of the replicon system, the one-plasmid system is easy to amplify and to deliver to the target cells with a regular transfection strategy, without a complex preparation process. The replicon with different reporter genes provided various options according to the requirement of individual researches. We designed multiple unique restriction sites in the replicon cDNA and made the replicon convenient for the manipulation of deletion, insertion and mutagenesis.
One advantage of the viral replicon system is its convenient usage in antiviral drug discovery. In this work, we transfected the cells with the reporter gene-carrying replicon and then treated the cells with various known drugs which inhibit the replication of SARS-CoV-2. Our data showed that the replicon with luciferase is sensitive to detect the antiviral effect of the candidate drugs as efficiently as live viruses (Fig. 4C–4I). Thus, our replicon is an efficient and convenient platform for antiviral drug screening.
In summary, we developed a biosafe and easy-to-use replicon system. Our data indicated that the replicon possessed full activity of the replication/transcription machinery and could produce a set of sgRNAs as the live virus. As the replicon is replication-competent but does not produce infectious virus, it may replace live virus in many assays in ordinary laboratories without causing concerns on biosafety. Thus, this biosafe replicon system is to benefit the scientific community in basic research and development of therapeutic strategies for SARS-CoV-2.
-
We thank Prof. Hui Zhang for providing the SYSU-1 strain of SARS-CoV-2 virus. This work was supported by Grants (the National Natural Science Foundation of China #32041002, #31971161, #31900546 and #81620108020); the Guangdong Science and Technology Department (#2019A1515011332); the Shenzhen Science and Technology Innovation Program (JSGG20200225150431472, JCYJ20190807160615255, JCYJ20190807153203560, and KQTD20180411143323605). D.G. is also supported by the Guangdong Zhujiang Leading Talents Programme and the National Tenthousand Talents Program.
-
XP, DG and JAP designed the experiments. YYJ, HL, LC, WCW, YJ, LD, YJ, YX, KT and FZ carried out the experiments. JAP, FX, MS and XP analyzed the data. XP, DG and JAP wrote the paper. DG checked and finalized the manuscript. All authors read and approved the final manuscript.
-
The authors declare that they have no conflict of interest.
-
This study was approved by the Ethics Committee of the Zhongnan Hospital, Wuhan University (No. 2020066). Informed consent was obtained from each patient or the immediate family. All sample processing and sequencing procedures were performed in accordance with the relevant guidelines and regulations of the biosafety facility of Zhongnan Hospital, Wuhan University, China.

















 DownLoad:
DownLoad: