-
Dear Editor,
Infectious bronchitis (IB), one of the most common and difficult poultry diseases, is caused by a gammacoronavirus named infectious bronchitis virus (IBV). IBV frequently causes respiratory and/or renal diseases in chickens and egg production losses in hens. IB has a global distribution (De Wit et al., 2011; Cook et al., 2012; Jackwood, 2012). In Morocco, IBV vaccine strains commercially available are of the Mass type (H120, Ma5, and modified Massachusetts strains), 793B type (4/91 and CR88), and (since 2013) Arkansas type. Because of its low price, H120 is the most commonly used vaccine in broiler chickens. Generally, IB has been controlled by serotype-specific vaccines, but outbreaks of IB still occur because vaccines do not always confer cross protection from serologically distinct virus variants.
The IBV genome consists of a single-stranded positive-sense RNA that encodes four structural proteins: spike (S), membrane (M), envelope (E), and nucleoprotein (N). New IBV serotypes and genotypes can emerge as a result of only very few mutations in amino-acid changes in the N-terminal part of the spike 1 (S1) subunit from nucleotide deletions, insertions, or point mutations (Cavanagh et al., 1992). Therefore, S1 gene sequencing and subsequent genetic analysis are used most frequently to determine the similarity of emerging IBV lineages and are important tools to monitor the phylogenetic and epidemiological evolution of IBV.
In Morocco, molecular and genetic characterization of IBV strain diversity have been very limited. A long-term retrospective study on Moroccan isolates is required to improve understanding of IBV evolution and IB epidemiology. The objective of this report is to perform a retrospective analysis of the origin and evolution of 62 Moroccan IBV isolates obtained from unvaccinated (21%) and vaccinated (70%) poultry flocks showing clinical signs of IB between 1983 and 2014.
IBV isolates were obtained from different IB outbreaks (Supplementary Table S1). All samples were inoculated into specific pathogen-free embryonated chicken eggs (Gelb Jr and Jackwood, 1998), and the harvested allantoic fluids were used to extract viral RNA using the MagMax Express semi-automatic extractor (Life Technologies, Grand Island, NY) according to the manufacturer’s instructions. Each sample of extracted RNA was used for real time reverse transcriptase-polymerase chain reaction (RT-PCR) amplification. For initial screening for IBV, we used real time RT-PCR with nucleoprotein (N) gene-specific oligonucleotides and probe as described previously (Meir et al., 2010).
The RT-PCR protocol and primers for amplification of the IBV S1 gene variable region (705–1,097 nucleotides, including the hypervariable region 3) were also previously described (Jones et al., 2005; Worthington et al., 2008). Reactions were performed in 25 µL volumes in a thermocycler (Applied Biosystems, Foster City, CA, USA; GeneAmp PCR system 9700). All primers used in this study are shown in Supplementary Table S2. Initial RT-PCR was performed using primers SX1+ and SX2–; secondary amplification (nested PCR) was performed using primers SX3+ and SX4–. RT-PCR products were purified using the Gene Clean Kit (ExoSAP-IT, Affymetrix, Santa Clara, CA, USA) and then sequenced in both directions using primers SX3+ and SX4–. Sanger sequencing was performed on a 16-capillary 3130XL genetic analyzer sequencer (Applied Biosystems).
To determine the phylogenetic relationships among IBV isolates, nucleotide sequences were aligned using open-source multiple sequence comparison by log expectation (MUSCLE), edited using Bioedit Software version 7.2.5 (Hall, 1999), and analyzed using open-source molecular evolutionary genetics analysis (MEGA) version 6.06 (Tamura et al., 2013). An ML phylogenetic tree was reconstructed, selecting as a substitution model that with the lowest Akaike information criterion: the Tamura 3-parameter model with gamma-distributed rates among sites. The robustness of the tree was established by bootstrap analysis with 1,000 replicates. Bootstrap values above 50 were labeled on major tree branches for reference to assign confidence levels to branches. Evolutionary distances between genotypes and within clusters of a genotype were calculated using the maximum composite likelihood distances in MEGA. The case histories of the studied Moroccan strains listed in (Supplementary Table S1), and nucleotide sequences were submitted to GenBank. A reference sequence was selected for each known (sub)lineage of IBV as recently described (Valastro et al., 2016).
The sequences of the 62 Moroccan IBV isolates have a nucleotide sequence identity between 39.9% (IBV/Morocco/97/1986 and IBV/Morocco/23/2013) and 100% (IBV/Morocco/18/2011 and IBV/Morocco/38/2011) compared to each other. The deduced amino-acid sequence identities of Moroccan IBV isolates ranged from 36.3% (IBV/Morocco/97/1986 and IBV/Morocco/ 23/2013) to 100% (IBV/Morocco/17/2013 and IBV/Morocco/10/2013).
Phylogenetic analysis revealed that one genotype (GI), including three lineages (GI-13, GI-21, and GI-1) has been present in Morocco over the last 31 years (Figure 1). Within the first lineage, GI-13 (so-called 793B, 4/91, or CR88), thirteen field strains formed four clusters represented by viruses isolated between 1983 and 2014. The clusters indicated in Figure 1 only aid the analysis of Moroccan IBV evolution but do not correspond to sub-genotypes.
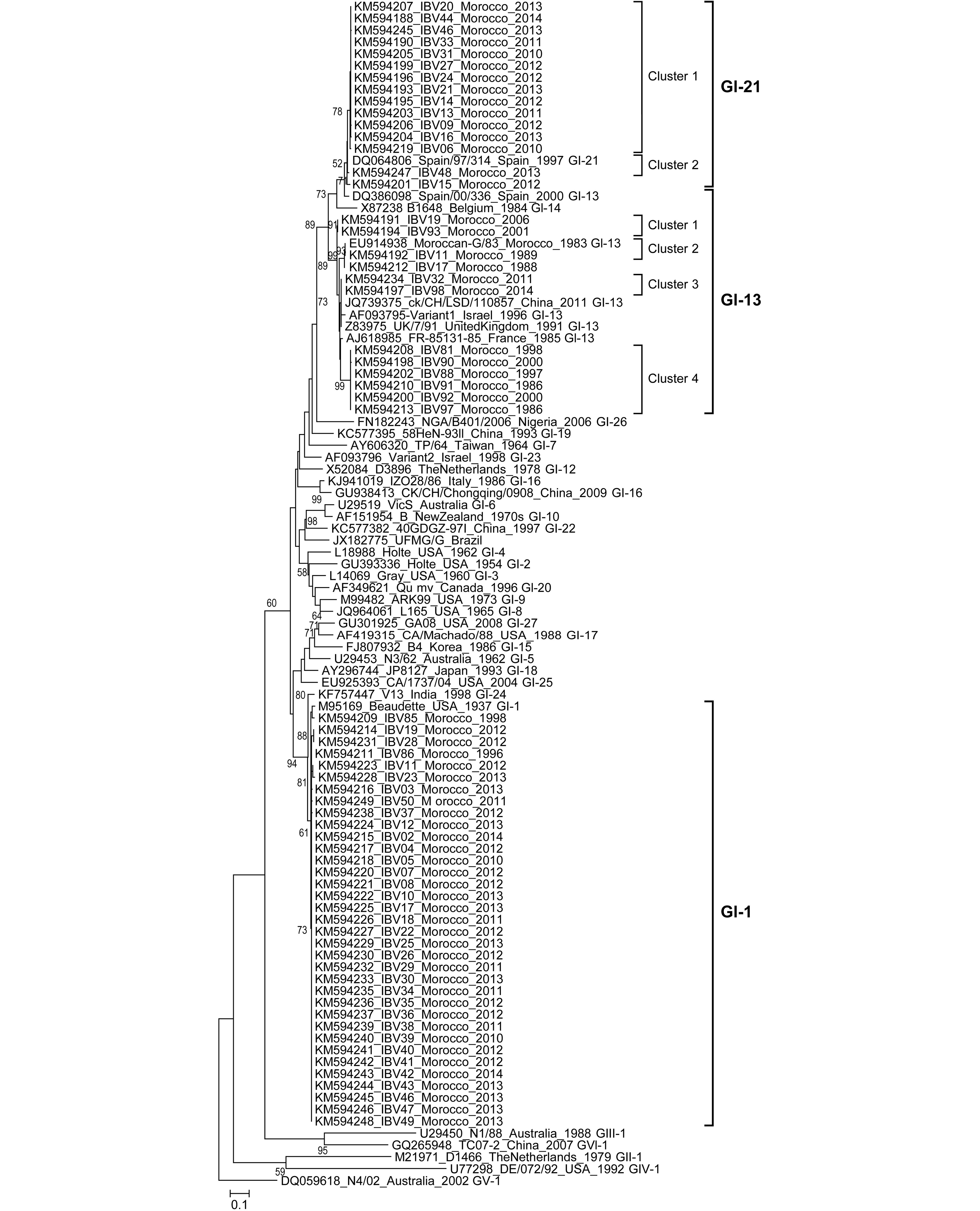
Figure 1. Phylogenetic tree representing the diversity of IBV strains in Morocco between 1983 and 2014. Bootstrap trials = 1,000. Bootstrap values above 50 were labeled on major tree branches for reference to assign confidence levels to branches. The last four digits of the sequence title represent the year of detection.
To study the degree of genetic diversity of IBV Moroccan strains, we analyzed the evolutionary distance between lineages. High genetic diversity was observed among three lineages with maximum likelihood (ML) distances ranging from 9.4% to 35.6% and 14.5% to 49.8% for nucleotide and amino-acid analyses, respectively (Supplementary Table S3). As a measure of the robustness of each node, we applied the ML method to a distance matrix obtained with bootstrap method (1,000 pseudo-replicas). The mean evolutionary distances within the three lineages, GI-1, -13, and -21, were 4.6%, 2.9%, and 0.1%, respectively.
The evolutionary distances comparisons for each lineage confirmed the clusters grouped by ML tree findings. The amino-acid sequence distances between the 4 clusters formed within the Moroccan IBV GI-13 lineage (previously called 793B) ranged from 5.2% to 11.6% (Supplementary Table S4). However, the methodology used to calculate genetic distances in the present study (maximum composite likelihood distances) presents a bias. The isolates from the GI-13 lineage had a nucleotide sequence identity ranging from 81.3% to 99.7%. Amino-acid sequence alignments revealed many point mutations and confirmed the four clusters within the GI-13 lineage (Supplementary Table S5). According to previous results, evidence suggests that GI-13 lineage evolved faster than Massachusetts lineage (GI-1) (Cavanagh et al., 1988; Adzhar et al., 1997).
Fifteen of the most recent Moroccan isolates were of the GI-21 lineage (Fellahi et al., 2015). Their nucleotide and deduced amino-acid sequence identities ranged from 82.7 to 100%. The maximum nucleotide and amino-acid sequence divergence within Moroccan GI-21 isolates were 5.3% and 6.3%, respectively. Phylogenetic analyses grouped the Moroccan GI-21 isolates into 2 clusters (Figure 1). The amino-acid sequence alignments revealed many point mutations and insertions in the partial S1 gene region (Supplementary Table S5). “Cluster 2” includes two Moroccan IBV GI-21 isolates and appears to have the closest phylogenetic relationship to the gCoV/AvCoV/chicken/Spain/1997 reference strain. “Cluster 1” includes only Moroccan IBV strains and has a very high percentage of identity, suggesting a common origin due to a single introduction. Morocco is the first African country in which the GI-21 lineage is reported. This lineage was among the most predominant lineages found across many countries in Europe (especially in Spain) and is spreading to other continents (Jones et al., 2005; Stooker L. Pan-European survey on the distribution of different strains of infectious bronchitis virus in 2011. Presented at the XVIII Congress of the World Veterinary Poultry Association, 19th-23rd August 2013, Nantes, France). This lineage could have been introduced into Morocco from Europe due to importation of 1-day-old broiler and layer breeders from Spain. Moreover, the high frequency of movement of personnel, vehicles, and other materials could act as vectors in the distribution of these viruses.
Fifty-five percent of the samples we characterized during the 2010–2014 period were of the GI-1 (Massachusetts-like) lineage (Fellahi et al., 2015). The nucleotide and deduced amino-acid sequence identities of these Moroccan IBV isolates were 99.1–100%. However, the Moroccan amino acid sequences were 100% identical with that of the IBV H120 (Massachusetts) vaccine strain (GenBank #M21970) used in the country. Globally, the GI-1 lineage comprises the first IBV serotype identified and even today is the most widely distributed genetic group, likely due to the extensive use of a homologous vaccine derived from one of its strains (Valastro et al., 2016).
This report is the first limited retrospective genotyping study of Moroccan IBV isolates. The heterogeneity between the numbers of samples per year is a bias in our retrospective study but is due to the small number of available archive strains in our laboratory. In addition, our analyses were based only on HVR3 of the S1 gene, which may bias IBV strain classification. Valastro et al. indeed recently recommended using the full S gene sequence to define IBV genotypes (Valastro et al., 2016), which, however, is not as straightforward to do in very resource-limited environments such as Moroccan laboratories.
As already observed in Italy (Franzo et al., 2014), vaccination plays an important role in the sustainability/disappearance of IBV genotypes. Evidence reported in this work confirms continuous evolution of IBV in poultry farms in Morocco and suggests that the distribution of IBV lineages should be carefully studied to implement appropriate vaccination programs.
-
We thank Laura Bollinger (NIH/NIAID Integrated Research Facility at Fort Detrick, Frederick, MD, USA) for critically editing the manuscript. This work was supported in part through Battelle Memorial Institute’s prime contract with the US National Institute of Allergy and Infectious Diseases (NIAID) under Contract No. HHSN272200700016I. A subcontractor to Battelle Memorial Institute who performed this work is: J.H.K., an employee of Tunnell Government Services, Inc. The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the US Department of Health and Human Services or of the institutions and companies affiliated with the authors. The authors declare that they have no conflict of interest. Tracheal or oropharyngeal swabs from live chickens suspected to be infected with IBV and from respiratory and renal tissues of dead chickens were collected with the approval of the Scientific Committee of the Agronomy and Veterinary Institute, Hassan II, Rabat, Morocco. Samplings was conducted under the guidelines of the Avian Pathology Unit and approved by the Office National de Sécurité Sanitaire des Aliments (ONSSA) under protocol n 25–08. All institutional and national guidelines for the care and use of laboratory animals were followed. Collected samples were then analyzed as described in this manuscript in the absence of further animal involvement, i.e. this article does not contain any studies with human participants or animals performed by any of the authors.
Supplementary tables are available on the Websites of Virologica Sinica: www.virosin.org;link.springer.com/journal/12250.
-
Codes IBV isolates Breed history reported Major clinical signs IBV vaccination Lineage GenBank accession numbers 1 Moroccan-G/83 Broiler Enteric N/A GI-13 EU914938 2 IBV/Morocco/97/1986 Broiler Respiratory Unvaccinated GI-13 KM594213 3 IBV/Morocco/91/1986 Broiler Respiratory Unvaccinated GI-13 KM594210 4 IBV/Morocco/17/1988 Broiler N/A N/A GI-13 KM594212 5 IBV/Morocco/11/1989 Broiler N/A N/A GI-13 KM594192 6 IBV/Morocco/86/1996 Broiler Respiratory Vaccinated GI-1 KM594211 7 IBV/Morocco/88/1997 Broiler Nephritis N/A GI-13 KM594202 8 IBV/Morocco/81/1998 Broiler Nephritis N/A GI-13 KM594208 9 IBV/Morocco/85/1998 Layers Respiratory Vaccinated GI-1 KM594209 10 IBV/Morocco/90/2000 Broiler Nephritis Vaccinated GI-13 KM594198 11 IBV/Morocco/92/2000 Broiler Nephritis Unvaccinated GI-13 KM594200 12 IBV/Morocco/93/2001 Broiler Nephritis N/A GI-13 KM594194 13 IBV/Morocco/19/2006 Broiler Nephritis Vaccinated GI-13 KM594191 14 IBV/Morocco/05/2010 Broiler Respiratory Vaccinated GI-1 KM594218 15 IBV/Morocco/06/2010 Broiler Respiratory Vaccinated GI-21 KM594219 16 IBV/Morocco/31/2010 Broiler Respiratory vaccinated GI-21 KM594205 17 IBV/Morocco/39/2010 Layer Respiratory vaccinated GI-1 KM594240 18 IBV/Morocco/13/2011 Broiler Nephritis Unvaccinated GI-21 KM594203 19 IBV/Morocco/18/2011 Broiler Respiratory vaccinated GI-1 KM594226 20 IBV/Morocco/29/2011 Broiler Respiratory vaccinated GI-1 KM594232 21 IBV/Morocco/32/2011 Broiler Nephritis vaccinated GI-13 KM594234 22 IBV/Morocco /33/2011 Broiler Nephritis vaccinated GI-21 KM594190 23 IBV/Morocco/34/2011 Breeder Respiratory vaccinated GI-1 KM594235 24 IBV/Morocco/38/2011 Layer Respiratory vaccinated GI-1 KM594239 25 IBV/Morocco/50/2011 Broiler Respiratory vaccinated GI-1 KM594249 26 IBV/Morocco/22/2012 Broiler Respiratory vaccinated GI-1 KM594227 27 IBV/Morocco/14/2012 Broiler Nephritis Unvaccinated GI-21 KM594195 28 IBV/Morocco/24/2012 Broiler Nephritis Unvaccinated GI-21 KM594196 29 IBV/Morocco/27/2012 Broiler Respiratory vaccinated GI-21 KM594199 30 IBV/Morocco/15/2012 Broiler Respiratory Unvaccinated GI-21 KM594201 31 IBV/Morocco/09/2012 Layer Respiratory vaccinated GI-21 KM594206 32 IBV/Morocco/19/2012 Broiler Respiratory vaccinated GI-1 KM594214 33 IBV/Morocco/04/2012 Broiler Respiratory vaccinated GI-1 KM594217 34 IBV/Morocco/07/2012 Broiler Respiratory vaccinated GI-1 KM594220 35 IBV/Morocco/08/2012 Broiler Nephritis vaccinated GI-1 KM594221 36 IBV/Morocco/11/2012 Layer Respiratory vaccinated GI-1 KM594223 37 IBV/Morocco/26/2012 Broiler Respiratory vaccinated GI-1 KM594230 38 IBV/Morocco/28/2012 Broiler Respiratory vaccinated GI-1 KM594231 39 IBV/Morocco/35/2012 Broiler Respiratory vaccinated GI-1 KM594236 40 IBV/Morocco/36/2012 Layer Respiratory vaccinated GI-1 KM594237 41 IBV/Morocco/37/2012 Broiler Respiratory vaccinated GI-1 KM594238 42 IBV/Morocco/40/2012 Broiler Respiratory vaccinated GI-1 KM594241 43 IBV/Morocco/41/2012 Broiler Respiratory vaccinated GI-1 KM594242 44 IBV/Morocco/21/2013 Broiler Respiratory Unvaccinated GI-21 KM594193 45 IBV/Morocco/16/2013 Broiler Respiratory vaccinated GI-21 KM594204 46 IBV/Morocco/20/2013 Broiler Nephritis vaccinated GI-21 KM594207 47 IBV/Morocco/03/2013 Broiler Respiratory vaccinated GI-1 KM594216 48 IBV/Morocco/10/2013 Broiler Respiratory vaccinated GI-1 KM594222 49 IBV/Morocco/12/2013 Broiler Respiratory vaccinated GI-1 KM594224 50 IBV/Morocco/17/2013 Broiler Respiratory Unvaccinated GI-1 KM594225 51 IBV/Morocco/23/2013 Broiler Respiratory vaccinated GI-1 KM594228 52 IBV/Morocco/25/2013 Broiler Respiratory vaccinated GI-1 KM594229 53 IBV/Morocco/30/2013 Broiler Respiratory vaccinated GI-1 KM594233 54 IBV/Morocco/43/2013 Broiler Respiratory Unvaccinated GI-1 KM594244 55 IBV/Morocco/46/2013 Broiler Respiratory Unvaccinated GI-21 KM594245 56 IBV/Morocco/47/2013 Broiler Respiratory Unvaccinated GI-1 KM594246 57 IBV/Morocco/48/2013 Broiler Respiratory Unvaccinated GI-21 KM594247 58 IBV/Morocco/49/2013 Broiler Respiratory vaccinated GI-1 KM594248 59 IBV/Morocco/02/2014 Broiler Respiratory vaccinated GI-1 KM594215 60 IBV/Morocco/98/2014 Broiler Nephritis vaccinated GI-13 KM594197 61 IBV/Morocco/42/2014 Broiler Respiratory vaccinated GI-1 KM594243 62 IBV/Morocco/44/2014 Broiler Respiratory vaccinated GI-21 KM594188 62 IBV/Morocco/45/2014 Broiler Nephritis vaccinated GI-1 KM594189 Note: The last four digits of the sequence title represent the year of detection. GI, genotype I; IBV, infectious bronchitis virus; N/A, data not available. Table S1. Data from 62 Moroccan IBV isolates included in this study
Primer name Primer sequence (5′–3′) Forward SX1+ CACCTAGAGGTTTGYTWGCAT Reverse SX2- TCCACCTCTATAAACACCYTT Forward SX3+ TAATACTGGYAATTTTTCAGA Reverse SX4- AATACAGATTGCTTACAACCACC Table S2. Primers used in this study
Genotype Amino acid evolutionary distance 1 2 3 4 5 6 7 8 9 10 11 1. GI-13 – 0.236 0.254 0.498 0.219 0.228 0.145 0.245 0.228 0.221 0.176 2. GI-9 0.170 0.227 0.483 0.220 0.194 0.208 0.257 0.231 0.262 0.257 3. GI-1 0.174 0.146 0.449 0.182 0.234 0.230 0.264 0.229 0.239 0.198 4. GII-1 0.356 0.349 0.318 0.481 0.491 0.469 0.483 0.501 0.468 0.496 5. GI-12 0.162 0.169 0.113 0.352 0.232 0.195 0.189 0.215 0.226 0.243 6. GI-3 0.169 0.129 0.137 0.364 0.165 0.205 0.269 0.242 0.269 0.248 7. GI-21 0.106 0.159 0.166 0.339 0.153 0.154 0.234 0.202 0.203 0.181 8. GI-26 0.173 0.181 0.176 0.349 0.150 0.183 0.169 0.266 0.227 0.266 9. GI-16 0.167 0.184 0.163 0.373 0.167 0.180 0.161 0.203 0.250 0.254 10. GI-19 0.171 0.197 0.157 0.365 0.177 0.201 0.158 0.179 0.193 0.267 11. GI-24 0.094 0.192 0.142 0.362 0.177 0178 0.126 0.192 0.185 0.188 – Nucleotide evolutionary distance Note: Shown are the base substitutions per site (evolutionary distance) from averaging over all sequence pairs between genetic groups. Analyses were conducted using the Maximum Composite Likelihood model. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). All positions containing gaps and missing data were eliminated. Table S3. Estimates of evolutionary distances over sequence pairs between genotypes
Genotype Amino acid evolutionary distance 1 2 3 4 5 Cluster 1 – 0.066 0.052 0.096 0.076 Cluster 2 0.051 0.068 0.116 0.090 Cluster 3 0.030 0.041 0.098 0.072 Cluster 4 0.063 0.084 0.060 0.089 GI-13 reference strains 0.050 0.066 0.044 0.056 – Nucleotide evolutionary distance Note: Shown are the base substitutions per site (evolutionary distance) from averaging over all sequence pairs between groups. Standard error estimate(s) are shown above the diagonal, in italic font. Analyses were conducted using the Maximum Composite Likelihood model. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). All positions containing gaps and missing data were eliminated. Table S4. Estimates of evolutionary distances over sequence pairs between Moroccan lineage GI-13 isolates
Strain Deletions or substitutions Insertions 268 271 285 287 289 293 296 297 299 301 306 326 329 338 340 345 364 291 351 GI-13 prototypea E I V N A S V D F Q H K D K N N I – – IBV/93/2001 E V V N A S V D F Q R K D P N N L – – IBV/19/2006 E V V N A S V D F Q R K D P N N L – – MoroccanG/83 E T V N S S V D F Q H I D E T T I – – IBV/11/1989 E T V N S S V D F Q H I D E T T I – – IBV/17/2013 E T V N S S V D F Q H I D E T T I – – IBV/98/2014 E T V N S S V D F Q H K D N N N I – – IBV/32/2011 E I V N A S V D F Q H K D K N N I – – IBV/81/1998 E I V N V L I E L V H K N P N N I – – IBV/90/2000 E I V N V L I E L V H K N P N N I – – IBV/88/1997 E I V N V L I E L V H K N P N N I – – IBV/91/1986 E I V N V L I E L V H K N P N N I – – IBV/92/2000 E I V N V L I E L V H K N P N N I – – IBV/97/1986 E I V N V L I E L V H K N P N N I – – GI-21 prototypeb E V E N P S V N F V H K D K N N I – – IBV/31/2010 E V V D P S V N F V H K D T N N I L W IBV/13/2011 E V V D P S V N F V H K D T N N I L W IBV/33/2011 E V V D P S V N F V H K D T N N I L W IBV/14/2012 E V V D P S V N F V H K D T N N I L W IBV/24/2012 E V V D P S V N F V H K D T N N I L W IBV/27/2012 E V V D P S V N F V H K D T N N I L W IBV/09/2012 E V V D P S V N F V H K D T N N I L W IBV/21/2013 E V V D P S V N F V H K D T N N I L W IBV/16/2013 E V V D P S V N F V H K D T N N I L W IBV/20/2013 E V V D P S V N F V H K D T N N I L W IBV/44/2014 E V V D P S V N F V H K D T N N I L W IBV/46/2013 E V V D P S V N F V H K D T N N I L W IBV/06/2010 E V V D P S V N F V H K D T N N I L W IBV/15/2012 E V E N P S V N F V H K D K N N I – – IBV/48/2013 E V E N P S V N F V H K D K N N I – – Note: aGI-13 prototype is gCoV/AvCoV/chicken/Spain/1989Spain/1997. bGI-21 prototype is gCoV/AvCoV/chicken/ Spain//97/314. Table S5. HVR3 S1 protein mutations of Moroccan IBV isolates from GI-13 and CG-21 lineages compared with reference prototypes
Phylogenetic analysis of avian infectious bronchitis virus isolates from Morocco: a retrospective study (1983 to 2014)
- Published Date: 12 April 2017
Abstract: In Morocco, molecular and genetic characterization of IBV strain diversity have been very limited. A long-term retrospective study on Moroccan isolates is required to improve understanding of IBV evolution and IB epidemiology. The objective of this report is to perform a retrospective analysis of the origin and evolution of 62 Moroccan IBV isolates obtained from unvaccinated (21%) and vaccinated (70%) poultry flocks showing clinical signs of IB between 1983 and 2014.
















 DownLoad:
DownLoad: