-
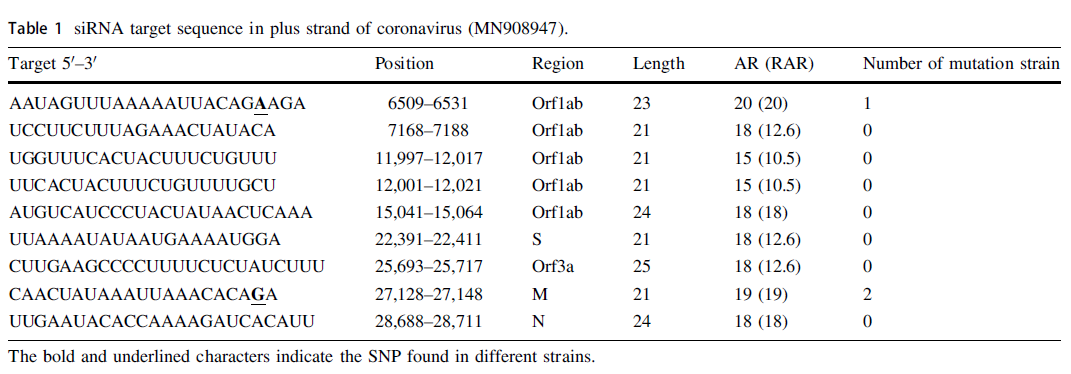
Computational Identification of Small Interfering RNA Targets in SARS-CoV-2
2020, 35(3): 359 doi: 10.1007/s12250-020-00221-6
Received: 06 March 2020 Accepted: 03 April 2020 Published: 15 April 2020With the epidemic of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) worldwide and in the absence of any effective vaccine, there is an urgent need to find a specific anti- SARS-CoV-2 agent. In this study, by analyzing the secondary structures of the SARS-CoV-2 genome (MN908947), several 21~25 base-long segments were obtained and selected as the potential targets of small interfering RNA duplexes. Moreover, it was also found that these targets are conserved among different strains. We hope the results will contribute to the pharmaceutical research and therapy of the SARS-CoV-2. -
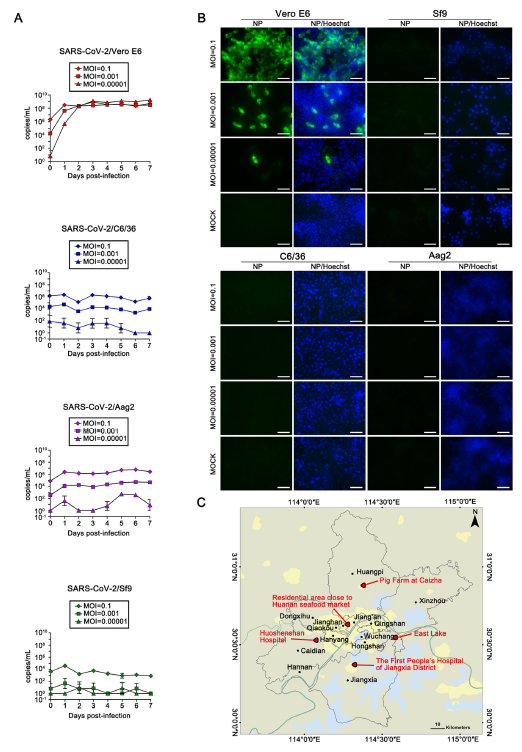
SARS-CoV-2 Does Not Replicate in Aedes Mosquito Cells nor Present in Field-Caught Mosquitoes from Wuhan
2020, 35(3): 355 doi: 10.1007/s12250-020-00251-0
Received: 10 May 2020 Accepted: 08 June 2020 Published: 29 June 2020With the rapid spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and growing fear, people have become very concerned about whether this novel coronavirus could be transmitted by mosquitoes. We evaluate the infectivity of SARS-CoV-2 in Aedes albopictus and Aedes aegypti derived cell lines. The results indicated that SARS-CoV-2 could not replicate in both C6/36, Sf9 and Aag2 cells. Further, no SARS-CoV-2 RNA was detected in the field collected Culex, and Anopheles mosquitoes during the months of April and May in Wuhan in 2020. Our findings highlight the restricted replication of SARS-CoV-2 in mosquito cells and field-caught mosquitoes. -
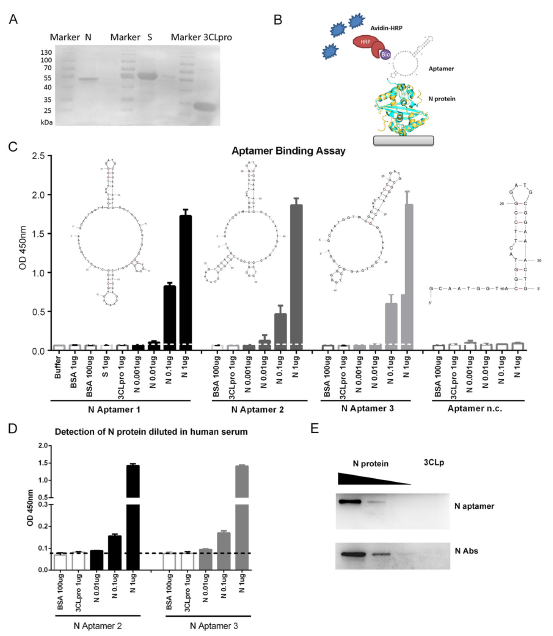
A DNA Aptamer Based Method for Detection of SARS-CoV-2 Nucleocapsid Protein
2020, 35(3): 351 doi: 10.1007/s12250-020-00236-z
Received: 16 April 2020 Accepted: 30 April 2020 Published: 25 May 2020Altogether, we have identified a novel method for the detection of SRAS-CoV-2 N protein using DNA based aptamers. Although the aptamers used in this study were designed based on an aptamer previously selected for SARS-CoV N protein, they bind to SRAS-CoV-2 N protein with a high affinity. Most importantly, SARS-CoV-2 N protein shares very low similarity (16%–38%) with N protein from other five known human coronaviruses except for SARS-CoV. Because there are no SARS-CoV cases reported since 2004, our aptamer-based method can be used as a supplementation to the current diagnosis of SARS-CoV-2. -
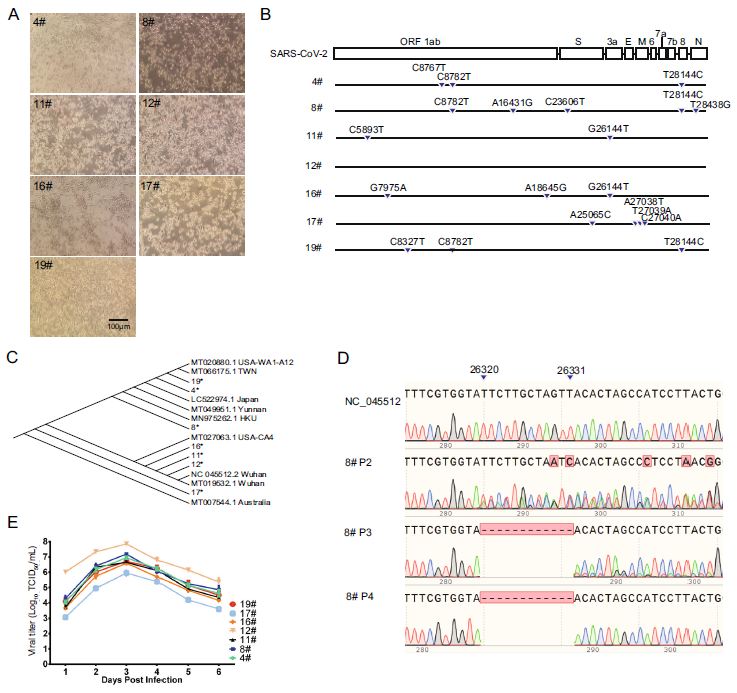
Isolation and Growth Characteristics of SARS-CoV-2 in Vero Cell
2020, 35(3): 348 doi: 10.1007/s12250-020-00241-2
Received: 01 April 2020 Accepted: 19 May 2020 Published: 19 June 2020The coronavirus disease 2019 (COVID-19) broke out in early December 2019 in Wuhan, China and escalated into a global pandemic. There is an urgent need to understand the biology of SARS-CoV-2. In this letter, we report the isolation and characterization of seven isolates of SARS-CoV-2. Results show that our viruses have 99% sequence identity with published virus sequences. In addition, all viruses grew well in Vero cells, and one of the viruses had a deletion mutation after short passage. These results shall facilitate the understanding of the characteristics of SARS-CoV-2 in vitro. -
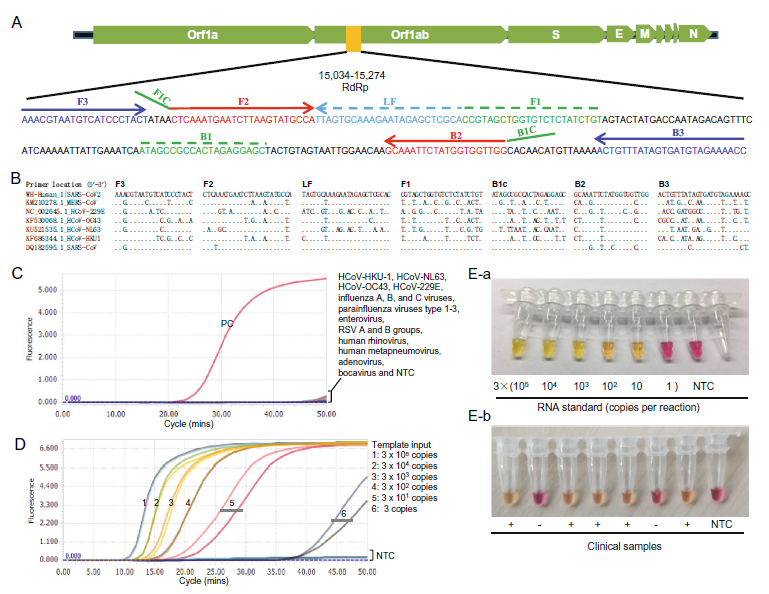
Development of a Novel Reverse Transcription Loop-Mediated Isothermal Amplification Method for Rapid Detection of SARS-CoV-2
2020, 35(3): 344 doi: 10.1007/s12250-020-00218-1
Received: 19 February 2020 Accepted: 22 March 2020 Published: 01 April 2020In conclusion, the novel visual RT-LAMP assay is a simple, rapid, and sensitive approach for detection of SARS-CoV-2, and it is ready for application in primary care and community hospitals or health care centers, and even patients' own houses in response to the current SARS-CoV-2 epidemic because the assay does not require sophisticated equipment and skilled personnel. Furthermore, it is also ready to be used in fields for screening samples from wild animals and environments to facilitate the identification of potential intermediate hosts that mediate the cross-species transmission of SARS-CoV-2 from bats to humans. -
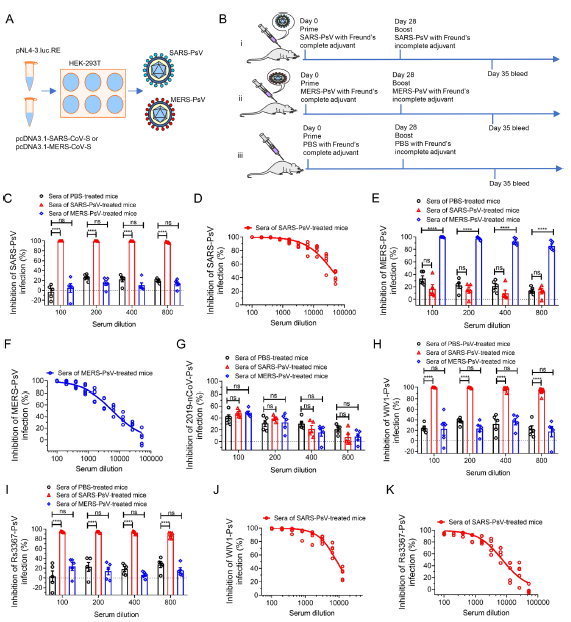
Inefficiency of Sera from Mice Treated with Pseudotyped SARS-CoV to Neutralize 2019-nCoV Infection
2020, 35(3): 340 doi: 10.1007/s12250-020-00214-5
Received: 22 February 2020 Accepted: 14 March 2020 Published: 31 March 2020The novel coronavirus 2019-nCoV has caused the pandemic of Wuhan pneumonia recently, posing a serious threat to global public health, and thus calling for the development of therapeutics and prophylactics. Here we showed that high titer anti-SARS-CoV spike protein serum cannot effectively neutralize 2019-nCoV infection. Based on our previous research, we developed SARS pseudovirus (SARS-PsV) and MERS pseudovirus (MERS-PsV) as immunogens to immunize mice. We found sera from mice treated with SARS-CoV S protein could potently cross-neutralize infection by SARS-CoV (50% neutralizing antibody titers, NT50 > 40, 000) and SARS-related coronavirus (NT50 > 7, 000), but weakly for 2019-nCoV infection NT50 < 100), implying that it may not be practical to treat 2019-nCoV infection with anti-SARS-CoV antibodies and that people with history of SARS-CoV infection many years ago may not be resistant to 2019-nCoV infection. -
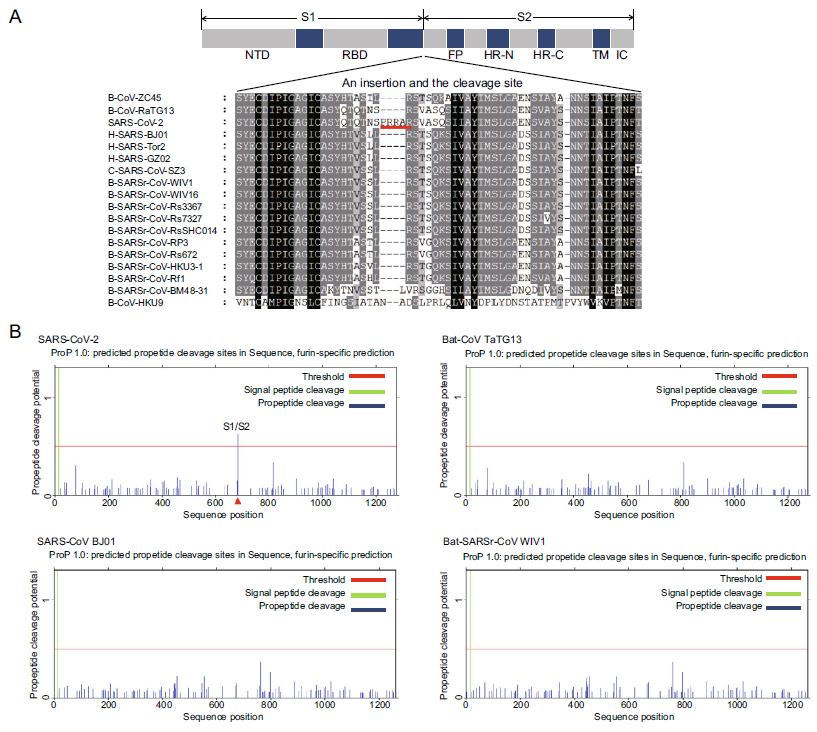
A Unique Protease Cleavage Site Predicted in the Spike Protein of the Novel Pneumonia Coronavirus (2019-nCoV) Potentially Related to Viral Transmissibility
2020, 35(3): 337 doi: 10.1007/s12250-020-00212-7
Received: 15 February 2020 Accepted: 06 March 2020 Published: 20 March 2020In summary, our sequence analysis on the S protein of 2019-nCoV has predicted a novel furin cleavage site at S1/S2 linkage. The ubiquitous expression of furin in different organs and tissues may be a reason for the high transmissibility and pathogenicity of 2019-nCoV observed in the current epidemic. However, since our findings were mainly based on bioinformatic analysis, more laboratory studies on 2019-nCoV in cell and animal models are required to verify our speculations and to avoid any bias. -
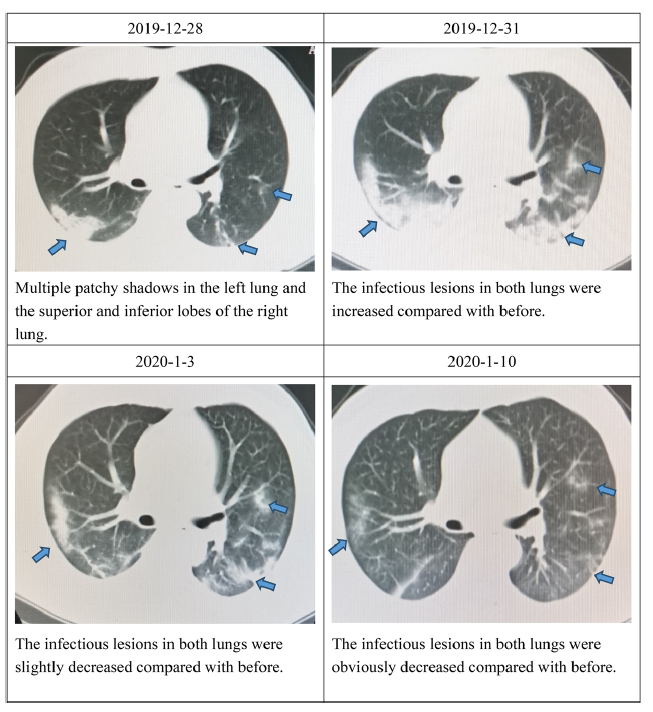
Clinical Features and Treatment of 2019-nCov Pneumonia Patients in Wuhan: Report of A Couple Cases
2020, 35(3): 330 doi: 10.1007/s12250-020-00203-8
Received: 25 January 2020 Accepted: 28 January 2020 Published: 07 February 2020Till January 20, 2020, the 2019-new coronavirus (2019-nCoV) has caused more than one hundred cases in Wuhan (WMHC 2020). During a retrospective study of recent pneumonia patients in our department, we found two patients who are likely being infected with the 2019-nCoV. During the hospitalization, those two patients were appropriately treated, and both were discharged within two weeks. Thus, we are reporting the clinical features and treatment regiment, and hope the information and experience can be shared. -
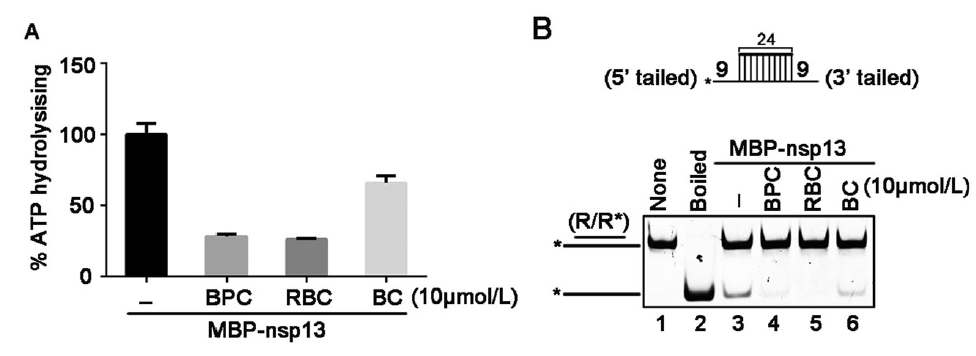
SARS-Coronavirus-2 Nsp13 Possesses NTPase and RNA Helicase Activities That Can Be Inhibited by Bismuth Salts
2020, 35(3): 321 doi: 10.1007/s12250-020-00242-1
Received: 07 May 2020 Accepted: 19 May 2020 Published: 04 June 2020The ongoing outbreak of Coronavirus Disease 2019 (COVID-19) has become a global public health emergency. SARS-coronavirus-2 (SARS-CoV-2), the causative pathogen of COVID-19, is a positive-sense single-stranded RNA virus belonging to the family Coronaviridae. For RNA viruses, virus-encoded RNA helicases have long been recognized to play pivotal roles during viral life cycles by facilitating the correct folding and replication of viral RNAs. Here, our studies show that SARS-CoV-2-encoded nonstructural protein 13 (nsp13) possesses the nucleoside triphosphate hydrolase (NTPase) and RNA helicase activities that can hydrolyze all types of NTPs and unwind RNA helices dependently of the presence of NTP, and further characterize the biochemical characteristics of these two enzymatic activities associated with SARS-CoV-2 nsp13. Moreover, we found that some bismuth salts could effectively inhibit both the NTPase and RNA helicase activities of SARS-CoV-2 nsp13 in a dose-dependent manner. Thus, our findings demonstrate the NTPase and helicase activities of SARS-CoV-2 nsp13, which may play an important role in SARS-CoV-2 replication and serve as a target for antivirals. -
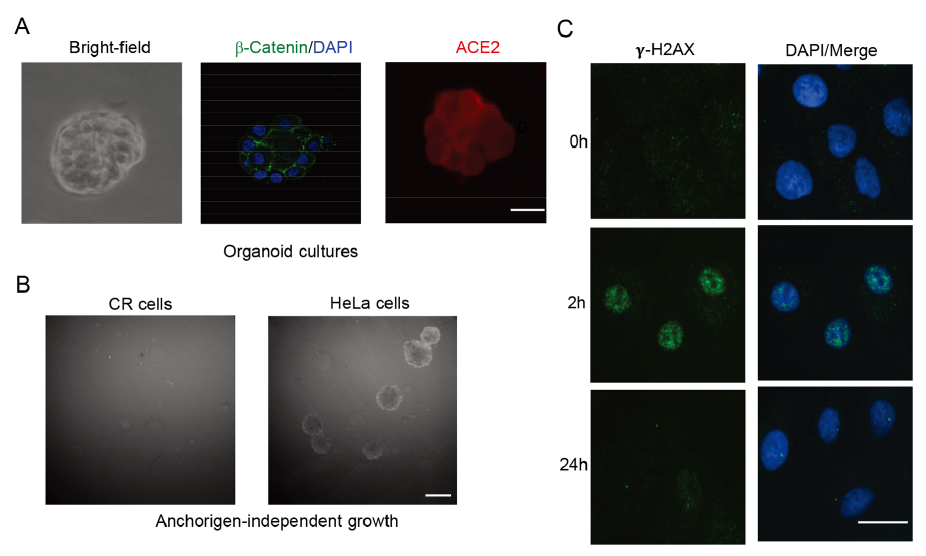
Long Term Culture of Human Kidney Proximal Tubule Epithelial Cells Maintains Lineage Functions and Serves as an Ex vivo Model for Coronavirus Associated Kidney Injury
2020, 35(3): 311 doi: 10.1007/s12250-020-00253-y
Received: 26 March 2020 Accepted: 09 June 2020 Published: 29 June 2020The mechanism of how SARS-CoV-2 causes severe multi-organ failure is largely unknown. Acute kidney injury (AKI) is one of the frequent organ damage in severe COVID-19 patients. Previous studies have shown that human renal tubule cells could be the potential host cells targeted by SARS-CoV-2. Traditional cancer cell lines or immortalized cell lines are genetically and phenotypically different from host cells. Animal models are widely used, but often fail to reflect a physiological and pathogenic status because of species tropisms. There is an unmet need for normal human epithelial cells for disease modeling. In this study, we successfully established long term cultures of normal human kidney proximal tubule epithelial cells (KPTECs) in 2D and 3D culture systems using conditional reprogramming (CR) and organoids techniques. These cells had the ability to differentiate and repair DNA damage, and showed no transforming property. Importantly, the CR KPTECs maintained lineage function with expression of specific transporters (SLC34A3 and cubilin). They also expressed angiotensin-converting enzyme 2 (ACE2), a receptor for SARS-CoV and SARS-CoV-2. In contrast, cancer cell line did not express endogenous SLC34A3, cubilin and ACE2. Very interestingly, ACE2 expression was around twofold higher in 3D organoids culture compared to that in 2D CR culture condition. Pseudovirion assays demonstrated that SARS-CoV spike (S) protein was able to enter CR cells with luciferase reporter. This integrated 2D CR and 3D organoid cultures provide a physiological ex vivo model to study kidney functions, innate immune response of kidney cells to viruses, and a novel platform for drug discovery and safety evaluation. -
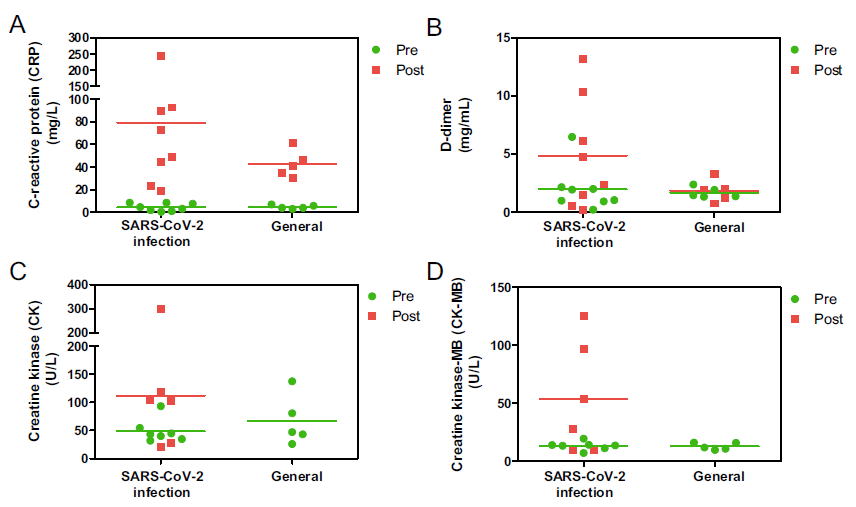
Clinical Manifestation and Laboratory Characteristics of SARS-CoV-2 Infection in Pregnant Women
2020, 35(3): 305 doi: 10.1007/s12250-020-00227-0
Received: 06 March 2020 Accepted: 10 April 2020 Published: 20 April 2020The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epidemic has become a major challenge to public health in China and other countries, considering its pathogenicity across all age groups. Pregnancy is a unique physiological condition, and is characterized by altered immunity and elevated hormone levels to actively tolerate the semi-allogeneic fetus, which undergoes a sudden and substantial fluctuation during the immediate postpartum period. Changes in clinical features, laboratory characteristics, and imaging features of pregnant women during the pre-partum and post-partum periods require further elucidation. Here, we retrospectively analyzed the clinical features, laboratory characteristics, and imaging features of eight pregnant cases of SARS-CoV-2 infection during the pre-partum and post-partum periods. Our results showed that four of the eight pregnant women were asymptomatic before delivery but became symptomatic post-partum. Correspondingly, white blood cell (WBC) counts increased and lymphocyte (LYMPH) counts decreased. C-reactive protein (CRP) levels in the serum also increased to a higher level than those in general pregnancy. Therefore, it is imperative to closely monitor laboratory parameters including the WBC count, LYMPH count, and CRP, along with other imaging features in chest CT scans, to promptly prevent, diagnose, and treat a SARS-CoV-2 infection during pregnancy. -
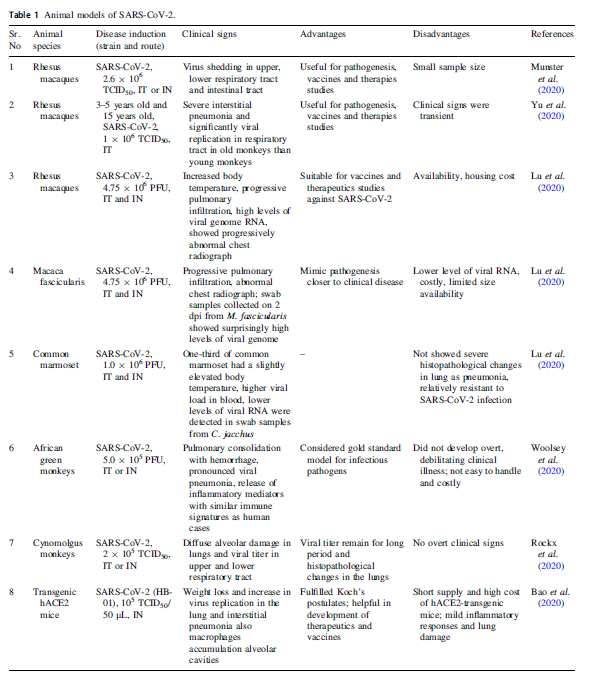
A Comprehensive Review of Animal Models for Coronaviruses: SARS-CoV-2, SARS-CoV, and MERS-CoV
2020, 35(3): 290 doi: 10.1007/s12250-020-00252-z
Received: 25 April 2020 Accepted: 02 June 2020 Published: 30 June 2020The recent outbreak of coronavirus disease (COVID-19) caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has already affected a large population of the world. SARS-CoV-2 belongs to the same family of severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV). COVID-19 has a complex pathology involving severe acute respiratory infection, hyper-immune response, and coagulopathy. At present, there is no therapeutic drug or vaccine approved for the disease. There is an urgent need for an ideal animal model that can reflect clinical symptoms and underlying etiopathogenesis similar to COVID-19 patients which can be further used for evaluation of underlying mechanisms, potential vaccines, and therapeutic strategies. The current review provides a paramount insight into the available animal models of SARS-CoV-2, SARS-CoV, and MERS-CoV for the management of the diseases. -
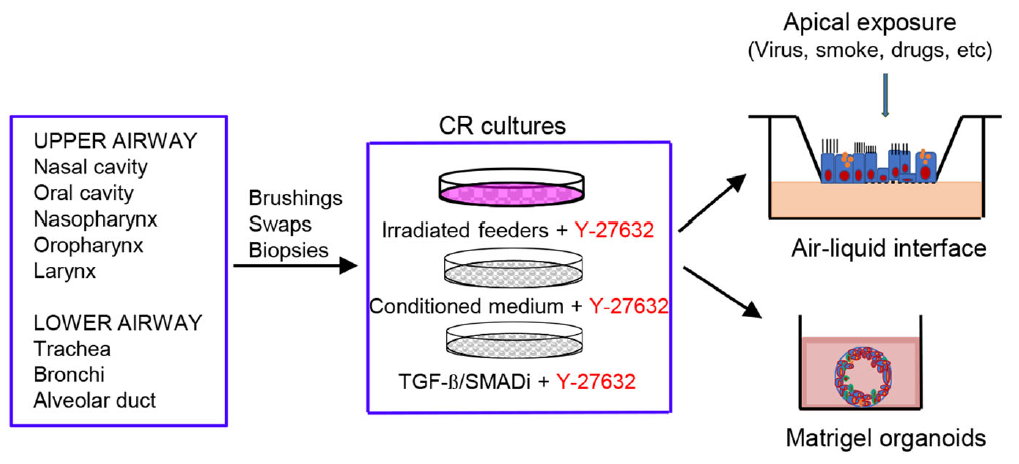
Conditionally Reprogrammed Human Normal Airway Epithelial Cells at ALI: A Physiological Model for Emerging Viruses
2020, 35(3): 280 doi: 10.1007/s12250-020-00244-z
Received: 16 April 2020 Accepted: 22 May 2020 Published: 17 June 2020Cancer cell lines have been used widely in cancer biology, and as biological or functional cell systems in many biomedical research fields. These cells are usually defective for many normal activities or functions due to significant genetic and epigenetic changes. Normal primary cell yields and viability from any original tissue specimens are usually relatively low or highly variable. These normal cells cease after a few passages or population doublings due to very limited proliferative capacity. Animal models (ferret, mouse, etc.) are often used to study virus-host interaction. However, viruses usually need to be adapted to the animals by several passages due to tropism restrictions including viral receptors and intracellular restrictions. Here we summarize applications of conditionally reprogrammed cells (CRCs), long-term cultures of normal airway epithelial cells from human nose to lung generated by conditional cell reprogramming (CR) technology, as an ex vivo model in studies of emerging viruses. CR allows to robustly propagate cells from non-invasive or minimally invasive specimens, for example, nasal or endobronchial brushing. This process is rapid (2 days) and conditional. The CRCs maintain their differentiation potential and lineage functions, and have been used for studies of adenovirus, rhinovirus, respiratory syncytial virus, influenza viruses, parvovirus, and SARS-CoV. The CRCs can be easily used for air-liquid interface (ALI) polarized 3D cultures, and these coupled CRC/ALI cultures mimic physiological conditions and are suitable for studies of viral entry including receptor binding and internalization, innate immune responses, viral replications, and drug discovery as an ex vivo model for emerging viruses. -
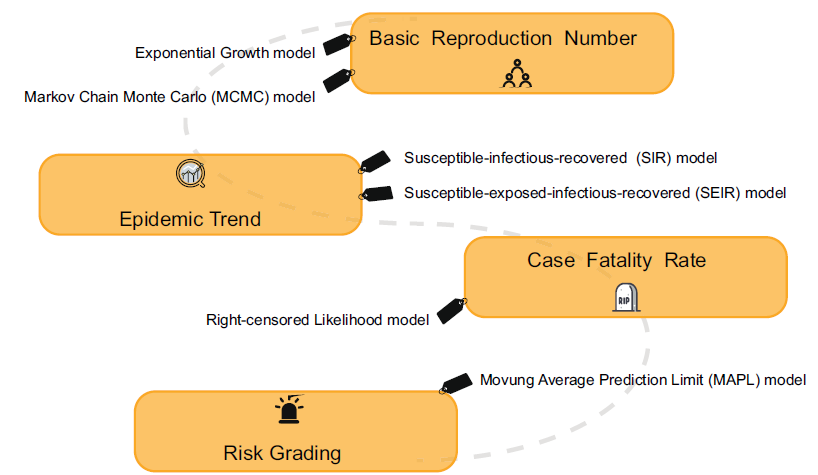
The Rapid Assessment and Early Warning Models for COVID-19
2020, 35(3): 272 doi: 10.1007/s12250-020-00219-0
Received: 08 March 2020 Accepted: 27 March 2020 Published: 01 April 2020Human beings have experienced a serious public health event as the new pneumonia (COVID-19), caused by the severe acute respiratory syndrome coronavirus has killed more than 3000 people in China, most of them elderly or people with underlying chronic diseases or immunosuppressed states. Rapid assessment and early warning are essential for outbreak analysis in response to serious public health events. This paper reviews the current model analysis methods and conclusions from both micro and macro perspectives. The establishment of a comprehensive assessment model, and the use of model analysis prediction, is very efficient for the early warning of infectious diseases. This would significantly improve global surveillance capacity, particularly in developing regions, and improve basic training in infectious diseases and molecular epidemiology. -
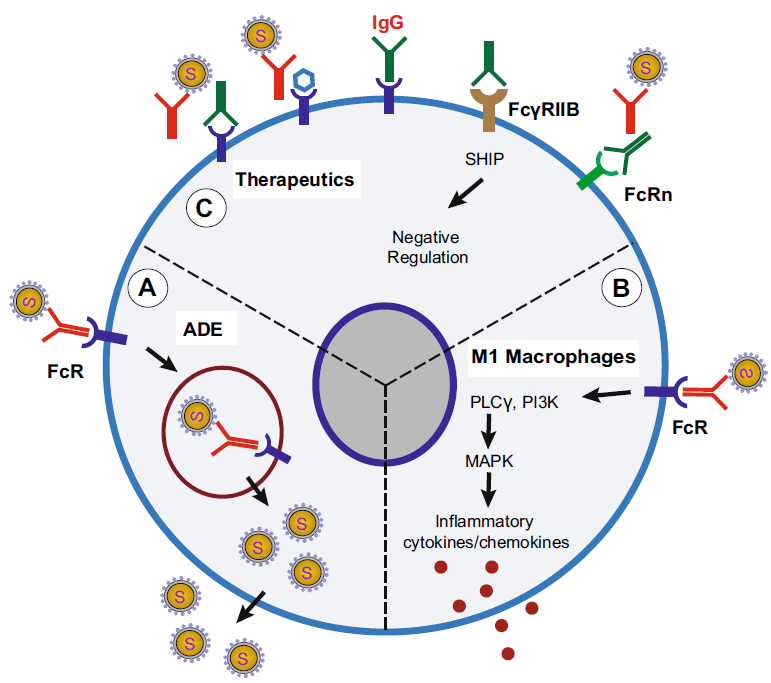
Understanding SARS-CoV-2-Mediated Inflammatory Responses: From Mechanisms to Potential Therapeutic Tools
2020, 35(3): 266 doi: 10.1007/s12250-020-00207-4
Received: 14 February 2020 Accepted: 16 February 2020 Published: 03 March 2020Currently there is no effective antiviral therapy for SARS-CoV-2 infection, which frequently leads to fatal inflammatory responses and acute lung injury. Here, we discuss the various mechanisms of SARS-CoV-mediated inflammation. We also assume that SARS-CoV-2 likely shares similar inflammatory responses. Potential therapeutic tools to reduce SARS-CoV-2-induced inflammatory responses include various methods to block FcR activation. In the absence of a proven clinical FcR blocker, the use of intravenous immunoglobulin to block FcR activation may be a viable option for the urgent treatment of pulmonary inflammation to prevent severe lung injury. Such treatment may also be combined with systemic anti-inflammatory drugs or corticosteroids. However, these strategies, as proposed here, remain to be clinically tested for effectiveness. -

The First Disease X is Caused by a Highly Transmissible Acute Respiratory Syndrome Coronavirus
2020, 35(3): 263 doi: 10.1007/s12250-020-00206-5
Received: 06 February 2020 Accepted: 09 February 2020 Published: 14 February 2020Based on the announcement of the World Health Organization (WHO) in 2018, the Wuhan pneumonia caused by an unknown etiology should be recognized as the first Disease X. Later, the pathogen was identified to be a novel coronavirus denoted 2019-nCoV, which has 79.5% and 96% whole genome sequence identify to SARS-CoV and bat SARS-related coronavirus (SARSr-CoV-RaTG13), respectively, suggesting its potential bat origin. With high human-to-human transmission rate (R0), 2019-nCoV has quickly spread in China and other countries, resulting in 34, 953 confirmed cases and 725 deaths as of 8 February 2020, thus calling for urgent development of therapeutics and prophylactics. Here we suggest renaming 2019-nCoV as "transmissible acute respiratory syndrome coronavirus (TARS-CoV)" and briefly review the advancement of research and development of neutralizing antibodies and vaccines targeting the receptor-binding domain (RBD) and viral fusion inhibitors targeting the heptad repeat 1 (HR1) domain in spike protein of 2019-nCoV. -
mRNA Vaccines: Possible Tools to Combat SARS-CoV-2
2020, 35(3): 259 doi: 10.1007/s12250-020-00243-0
Received: 20 March 2020 Accepted: 18 May 2020 Published: 10 June 2020Since December 2019, a novel coronavirus pneumonia outbreak has been reported in 212 countries and territories, with over 5, 000, 000 infections and over 300, 000 deaths on May 22, 2020. To date, except some repurposed drugs have been shown inhibitory effect on the so-called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), no drugs have been licensed to be effective and safe in treating COVID-19. Inspite of emergent need, no authorized vaccines have been approved for clinical use against this new virus currently. After the genetic map of SARS-CoV-2 was shared, scientists and biotech and vaccine companies promptly launched a race to pursue different types of vaccines to prevent illness from this novel virus. Among these vaccine projects, mRNA vaccines are showing a promising future with their feature of fast development. -
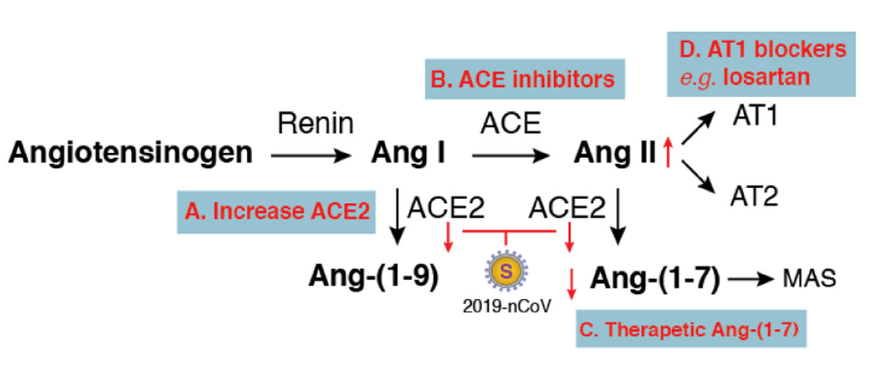
Compensation of ACE2 Function for Possible Clinical Management of 2019-nCoV-Induced Acute Lung Injury
2020, 35(3): 256 doi: 10.1007/s12250-020-00205-6
Received: 01 February 2020 Accepted: 03 February 2020 Published: 07 February 2020This perspective is intended to stimulate discussions about possible pathogenic mechanisms, based on the interaction of the virus with its receptor ACE2, so that rational therapies can be developed. Although ACE is regarded as the primary Ang II-converting enzyme, another enzyme, chymase, is also involved in converting Ang II in certain pathological conditions that may need attention (Fyhrquist and Saijonmaa 2008; Lindberg et al. 1997). In addition, coronavirus pathogenesis is a highly complex process (Lo et al. 2006), and much of the needed detail in host–pathogen interaction in 2019-nCoV infection awaits investigation. It also remains to be clinically tested whether some of these potential treatments, as proposed here, can be effective or beneficial in the management of 2019-nCoV-induced lung injury. -

Old Weapon for New Enemy: Drug Repurposing for Treatment of Newly Emerging Viral Diseases
2020, 35(3): 253 doi: 10.1007/s12250-020-00204-7
Received: 29 January 2020 Accepted: 31 January 2020 Published: 11 February 2020In December 2019, a dozen of patients with unusual pneumonia were hospitalized in Wuhan in central China, and the causative agent was identified as a new type of coronavirus (Zhu et al. 2020; Huang et al. 2020). The new virus was temporarily named as 2019 novel coronavirus (2019-nCoV) by the World Health Organization (WHO). As of January 29, 2020, 7736 confirmed cases of 2019-nCoV infection with 170 deaths were reported in China, and additional 77 cases in other 16 countries (National Health Commission of the People's Republic of China 2020; WHO 2020c). Since the emerging viruses are previously unknown pathogens, there are no specific and effective drugs available. Therefore, there is an urgent need for antiviral treatment in fighting the emerging viral diseases. However, the development of antiviral drugs is time- and resource-consuming, and thus repurposing of existing drugs to treat emerging viral diseases represents one of efficient strategies for drug development.


















