
THE THREE G's Meeting the Invertebrate Cell Culture Pioneers: Goldschmidt, Gaw, and Grace
2012, 27(6): 325 doi: 10.1007/s12250-012-3295-0
Received: 14 November 2012
Accepted: 22 November 2012
It was my good fortune to meet personally the three invertebrate cell culture pioneers, Richard Goldschmidt, Zan-Yin Gaw, and Thomas D. C. Grace (Fig. 1). In 1951 I met Goldschmidt at a symposium in Cold Spring Harbor, but I only knew that he was a prominent geneticist. I had no idea about his insect cell culture work at Yale University and daily contacts with Ross G. Harrison. In 1959 Zan Yin Gaw in Wuhan successfully cultured monolayers of silkworm cells for more than one year. I reported his breakthrough achievement at the 11th International Congress of Entomology in Vienna in 1960, but his work was completely ignored outside China. In 1982 Gaw invited me to Wuhan where he told me that he studied in the United States in the 1930s, working as postdoctoral scientist at the Rockefeller Institute, where he was daily meeting William Trager, and later at Yale University in the Osborn Laboratory, where he was inspired by Harrison. T. D. C. Grace worked in my laboratory at Rockefeller University during 1957 and 1958, then returned to CSIRO in Canberra, Australia. In 1962 he successfully established a cell line from the ovarian tissues of Antherea eucalypti pupae. The subsequent expansion of invertebrate cell culture involved the Nobel laureate Rita Levi Montalcini. I met her at a Growth Symposium, where I saw Harrison the last time. In 1969 she published the first of a dozen papers on in vitro studies of the embryonic nerve system of Periplaneta americana that led to her milestone discovery of the nerve growth factors. The “3 G-s” insect cell culture pioneers were directly or indirectly influenced and inspired by Harrison. Gaw was the only one who created a large following of insect cell culture researchers, who continued and expanded the work started by him in China.
The miRNAs of Herpes Simplex Virus (HSV)*
2012, 27(6): 333 doi: 10.1007/s12250-012-3266-5
Received: 28 June 2012
Accepted: 24 October 2012
Herpes simplex virus (HSV) is a group of common human pathogens with two serotypes HSV-1 and HSV-2. The prevalence of HSV is worldwide. It primarily infects humans through epithelial cells, when it introduces a latent infection into the nervous system. During viral latency, only a region known as the latency-associated transcript (LAT) is expressed. The discovery of HSV miRNAs helps to draw a larger picture of the infection and pathogenesis of the virus. This review summarizes miRNAs found in HSV-1 and HSV-2 so far. The functional studies of miRNAs in HSV to date indicate that they play a stage-specific role coordinated with viral proteins to maintain the virus life cycle.
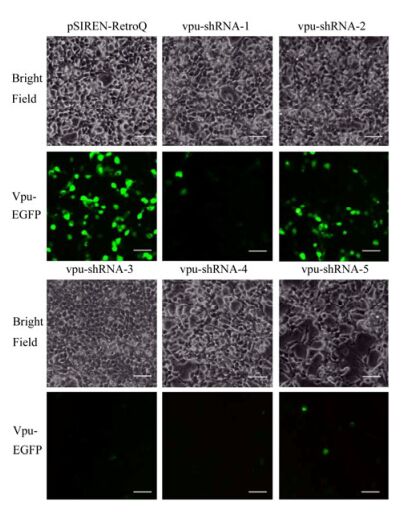
Two Retroviruses Packaged in One Cell Line can Combined Inhibit the Replication of HIV-1 in TZM-bl Cells *
2012, 27(6): 339 doi: 10.1007/s12250-012-3263-8
Received: 13 June 2012
Accepted: 07 November 2012
The cellular protein tetherin tethers the HIV-1 viral particles on the cellular membrane to inhibit the replication of HIV-1. However, the HIV-1 accessory protein Vpu counteracts the antiviral function of tetherin. In this study, two retroviral vector plasmids were constructed. One inhibited the vpu gene expression; the other one over-expressed the tetherin. Both retroviral vector plasmids could be packaged in the packaging cell line PT67 to obtain the corresponding retroviruses. The retroviral vector plasmids' functions of tetherin over-expression or vpu-RNAi were detected at the cell level. Retroviral vector plasmids were transfected to PT67 cells at different ratios from 0T3V to 3T0V, and then mixed retroviruses were harvested. The antiviral functions of mixed retroviruses were detected in HIV-1 infected TZM-bl cells. The results showed that packaged mixed retroviruses could repress the replication of HIV-1 in TZM-bl cells.
Immunogenic and Protective Properties of the First Kazakhstan Vaccine against Pandemic Influenza A(H1N1) pdm09 in Ferrets
2012, 27(6): 345 doi: 10.1007/s12250-012-3272-7
Received: 18 July 2012
Accepted: 16 October 2012
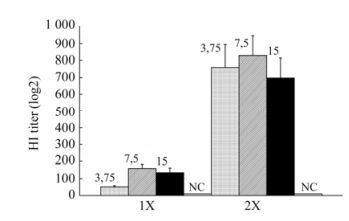
This paper presents the results of a pre-clinical study of the immunogenicity and efficacy of an egg-derived, inactivated, whole-virion adjuvanted vaccine (Refluvac?) on ferret models. For this purpose, groups of eight ferrets (6 to 7 months old) were injected with 0.5 mL of vaccine specimens containing 3.75, 7.5 or 15.0 μg of virus hemagglutinin. Administration was intramuscular and given either as a single dose or as two doses 14 days apart. All vaccine specimens manifested immunogenicity in ferrets for single (HI titer, from 51 ± 7 to 160 ± 23) and double (HI titer, from 697 ± 120 to 829 ± 117) administrations. To assess the protective effects of the vaccine, ferrets from the vaccinated and control groups were infected intranasally with pandemic virus А/California/7/09 (Н1N1) pdm09 at a dose of 106 EID50/0.5 mL. Fourteen days post-infection, the ferrets inoculated with single or double vaccines containing 3.75, 7.5 or 15.0 μg of hemagglutinin per dose showed no signs of influenza infection, weight loss, or body temperature rise, and no premature deaths occurred. The number of vaccinated ferrets shedding the virus via the upper airway, as well as the amount of virus shed after infection, was significantly reduced in comparison with animals from the control group. Based on our results, we sug-gest that a single vaccination at a dose of 3.75 or 7.5 μg hemagglutinin be used for Phase I clinical trials.
Differentiation of Sheeppox and Goatpox Viruses by Polymerase Chain Reaction-Restriction Fragment Length Polymorphism
2012, 27(6): 353 doi: 10.1007/s12250-012-3277-2
Received: 28 July 2012
Accepted: 29 October 2012
In the present study, the partial gene sequences of P32 protein, an immunogenic envelope protein of Capripoxviruses (CaPV), were analyzed to assess the genetic relationship among sheeppox and goatpox virus isolates, and restriction enzyme specific PCR-RFLP was developed to differentiate CaPV strains. A total of six goatpox virus (GTPV) and nine sheeppox virus (SPPV) isolates of Indian origin were included in the sequence analysis of the attachment gene. The sequence analysis revealed a high degree of sequence identity among all the Indian SPPV and GTPV isolates at both nucleotide and amino acid levels. Phylogenetic analysis showed three distinct clusters of SPPV, GTPV and Lumpy skin disease virus (LSDV) isolates. Further, multiple sequence alignment revealed a unique change at G120A in all GTPV isolates resulting in the formation of Dra I restriction site in lieu of EcoR I, which is present in SPPV isolates studied. This change was unique and exploited to develop restriction enzyme specific PCR-RFLP for detection and differentiation of SPPV and GTPV strains. The optimized PCR-RFLP was validated using a total of fourteen (n=14) cell culture isolates and twenty two (n=22) known clinical samples of CaPV. The Restriction Enzyme specific PCR-RFLP to differentiate both species will allow a rapid differential diagnosis during CaPV outbreaks particularly in mixed flocks of sheep and goats and could be an adjunct/supportive tool for complete gene or virus genome sequencing methods.
Specific VpU Codon Changes were Significantly associated with gp120 V3 Tropic Signatures in HIV-1 B-subtype
2012, 27(6): 360 doi: 10.1007/s12250-012-3287-0
Received: 17 September 2012
Accepted: 24 October 2012
After infection and integration steps, HIV-1 transcriptions increase sharply and singly-spliced mRNAs are produced. These encode Env (gp120 and gp41) and auxiliary proteins Vif, Vpr and VpU. The same localization within the unique structure of the mRNAs suggests that the VpU sequence prior to the Env could affect the Env polyprotein expression. The HIV-1 infection process begins when the gp120 subunit of the envelope glycoprotein complex interacts with its receptor(s) on the target cell. The V3 domain of gp120 is the major determinant of cellular co-receptor binding. According to phenotypic information of HIV-1 isolates, sequences from the VpU to V3 regions (119 in R5- and 120 X4-tropic viruses; one per patient) were analysed. The binomial correlation phi coefficient was used to assess covariation among VpU and gp120V3 signatures. Subsequently, average linkage hierarchical agglomerative clustering was performed. Beyond the classical V3 signatures (R5-viruses: S11, E25D; X4-viruses: S11KR, E25KRQ), other specific V3 and novel VpU signatures were found to be statistically associated with co-receptor usage. Several statistically significant associations between V3 and VpU mutations were also observed. The dendrogram showed two distinct large clusters: one associated with R5-tropic sequences (bootstrap=0.94), involving: (a) H13NPV3, E25DV3, S11V3, T22AV3 and Q61HVpU, (b) E25AV3 and L12FVpU, (c) D44EVpU, R18QV3 and D80NVpU; and another associated with X4-tropic sequences (bootstrap=0.97), involving: (i) E25IV3 and V10AVpU, (ii) 0-1insVVpU, H13RV3, I46LVpU, I30MV3 and 60-62delVpU, (iii) S11KRV3 and E25KRQV3. Some of these pairs of mutations were encoded always by one specific codon. These data indicate the possible VpU mutational patterns contributing to regulation of HIV-1 tropism.
A Case of Hepatitis B Reactivation due to the Hepatitis B Virus Escape Mutant in a Patient undergoing Chemotherapy *
2012, 27(6): 369 doi: 10.1007/s12250-012-3284-3
Received: 11 September 2012
Accepted: 05 November 2012
A 62-year-old man had chronic hepatitis B virus (HBV) infection and was diagnosed with liver cirrhosis. At the time of diagnosis the patient’s virologic markers were positive for hepatitis B surface antigen (HBsAg), antibody to hepatitis B e antigen (anti-HBe) and antibody to hepatitis B core antigen (anti-HBc), while antibody to hepatitis B surface antigen (anti-HBs) and HBV DNA were negative. Later the patient received chemotherapy for malignancy. However, this was interrupted due to elevated liver enzymes. At the same time HBV DNA became positive. Lamivudine (LMV) therapy was administered immediately. However, the levels of serum aminotransferase and total bilirubin (TB) were still rising. Finally the patient died of fulminant hepatic failure. A sequence revealed HBV genotype C (HBsAg subtype adw) with immune escape mutations, F8L, S34L, F41S, G44V, F93C, V96G, L110I, C149Y and F161Y. The high morbidity and mortality of this complication is one of the major obstacles to completing the standard treatment for malignancy in HBV carriers. Therefore, the relative risk of antiviral prophylactic failure should be further assessed and the the optimal strategy for antiviral prophylaxis in HBsAg-positive patients with oncologic and hematologic malignancies undergoing chemotherapy should be revised.
An Effective Mannosylated Chitosan Nanoparticle DNA Vaccine for FMD Virus
2012, 27(6): 373 doi: 10.1007/s12250-012-3269-2
Received: 05 July 2012
Accepted: 07 November 2012
We are reporting you an effective DNA vaccine for FMD virus, and it was prepared using mannosylated chitosan (MC) nanoparticles to form the FMDV-pVAC-VP1-OmpA complex particles. These particles were characterized for their physical properties like morphology, size and charge prior evaluating the actual vaccine delivery effect of MC-nanoparticles. The immunological evaluation indicated that the 20 μg of DNA vaccine complexed with MC nano-particles was found optimum in inducing the immune response in G pigs as measured by FMDV specific neutralising antibodies and Th1/Th2 responses using micro SNT and ELISA, respectively.


















