Advances of Bioinformatics Tools Applied in Virus Epitopes Prediction
2011, 26(1): 1 doi: 10.1007/s12250-011-3159-4
Received: 19 July 2010
Accepted: 16 October 2010
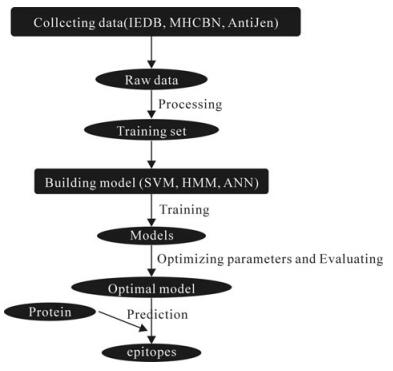
In recent years, the in silico epitopes prediction tools have facilitated the progress of vaccines development significantly and many have been applied to predict epitopes in viruses successfully. Herein, a general overview of different tools currently available, including T cell and B cell epitopes prediction tools, is presented. And the principles of different prediction algorithms are reviewed briefly. Finally, several examples are present to illustrate the application of the prediction tools.
Production and Characterization of Monoclonal Antibodies to Bluetongue Virus
2011, 26(1): 8 doi: 10.1007/s12250-011-3171-8
Received: 16 October 2010
Accepted: 10 December 2010
In the present study, a total of 24 MAbs were produced against bluetongue virus (BTV) by polyethyleneglycol (PEG) mediated fusion method using sensitized lymphocytes and myeloma cells. All these clones were characterized for their reactivity to whole virus and recombinant BTV-VP7 protein, titres, isotypes and their reactivity with 24 BTV-serotype specific sera in cELISA. Out of 24 clones, a majority of them (n = 18) belong to various IgG subclasses and the remaining (n = 6) to the IgM class. A panel of eight clones reactive to both whole BTV and purified rVP7 protein were identified based on their reactivity in iELISA. For competitive ELISA, the clone designated as 4A10 showed better inhibition to hyperimmune serum of BTV serotype 23. However, this clone showed a variable percent of inhibition ranging from16.6% with BTV 12 serotype to 78.9% with BTV16 serotype using 24 serotype specific sera of BTV originating from guinea pig at their lowest dilutions. From the available panel of clones, only 4A10 was found to have a possible diagnostic application.
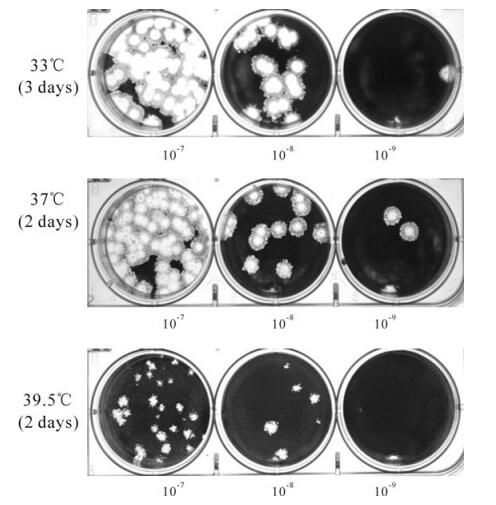
Construction and Genetic Analysis of Murine Hepatitis Virus Strain A59 Nsp16 Temperature Sensitive Mutant and the Revertant Virus
2011, 26(1): 19 doi: 10.1007/s12250-011-3145-x
Received: 23 April 2010
Accepted: 19 November 2010
Coronaviruses (CoVs) are generally associated with respiratory and enteric infections and have long been recognized as important pathogens of livestock and companion animals. Mouse hepatitis virus (MHV) is a widely studied model system for Coronavirus replication and pathogenesis. In this study, we created a MHV-A59 temperature sensitive (ts) mutant Wu”-ts18(cd) using the recombinant vaccinia reverse genetics system. Virus replication assay in 17C1-1 cells showed the plaque phenotype and replication characterization of constructed Wu”-ts18(cd) were indistinguishable from the reported ts mutant Wu”-ts18. Then we cultured the ts mutant Wu”-ts18(cd) at non-permissive temperature 39.5°C, which “forced” the ts recombinant virus to use second-site mutation to revert from a ts to a non-ts phenotype. Sequence analysis showed most of the revertants had the same single amino acid mutation at Nsp16 position 43. The single amino acid mutation at Nsp16 position 76 or position 130 could also revert the ts mutant Wu”-ts18 (cd) to non-ts phenotype, an additional independent mutation in Nsp13 position 115 played an important role on plaque size. The results provided us with genetic information on the functional determinants of Nsp16. This allowed us to build up a more reasonable model of CoVs replication-transcription complex.
A Simple and Rapid Colloidal Gold-based Immunochromatogarpic Strip Test for Detection of FMDV Serotype A
2011, 26(1): 30 doi: 10.1007/s12250-011-3166-5
Received: 24 August 2010
Accepted: 08 December 2010
A sandwich format immunochromatographic assay for detecting foot-and-mouth disease virus (FMDV) serotypes was developed. In this rapid test, affinity purified polyclonal antibodies from Guinea pigs which were immunized with sucking-mouse adapted FMD virus (A/AV88(L) strain) were conjugated to colloidal gold beads and used as the capture antibody, and affinity purified polyclonal antibodies from rabbits which were immunized with cell-culture adapted FMD virus (A/CHA/09 strain) were used as detector antibody. On the nitrocellulose membrane of the immunochromatographic strip, the capture antibody was laid on a sample pad, the detector antibody was printed at the test line(T) and goat anti-guinea pigs IgG antibodies were immobilized to the control line(C). The lower detection limit of the test for a FMDV 146S antigen is 11.7ng/ml as determined in serial tests after the strip device was assembled and the assay condition optimization. No cross reactions were found with FMDV serotype C, Swine vesicular disease (SVD), Vesicular stomatiti svirus (VSV) and vesicular exanthema of swine virus (VES) viral antigens with this rapid test. Clinically, the diagnostic sensitivity of this test for FMDV serotypes A was 88.7% which is as same as an indirect-sandwich ELISA. The specificity of this strip test was 98.2% and is comparable to the 98.7% obtained with indirect-sandwich ELISA. This rapid strip test is simple, easy and fast for clinical testing on field sites; no special instruments and skills are required, and the result can be obtained within 15 min. To our knowledge, this is the first rapid immunochromatogarpic assay for serotype A of FMDV
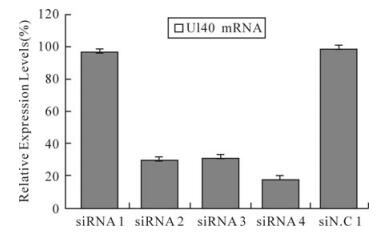
Effect of siRNAs on HSV-1 Plaque Formation and Relative Expression Levels of RR mRNA
2011, 26(1): 40 doi: 10.1007/s12250-011-3162-9
Received: 03 August 2010
Accepted: 30 September 2010
RNA interference (RNAi) is a process by which introduced small interfering RNA (siRNA) can cause the specific degradation of mRNA with identical sequences. The human herpes simplex virus type 1 (HSV-1) RR is composed of two distinct homodimeric subunits encoded by UL39 and UL40, respectively. In this study, we applied siRNAs targeting the UL39 and UL40 genes of HSV-1. We showed that synthetic siRNA silenced effectively and specifically UL39 and UL40 mRNA expression and inhibited HSV-1 replication. Our work offers new possibilities for RNAi as a genetic tool for inhibition of HSV-1 replication
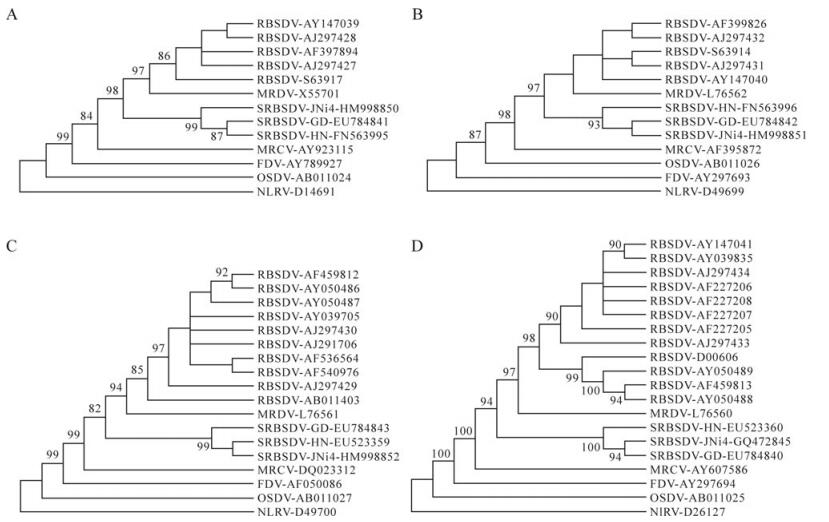
Molecular Characterization of Segments S7 to S10 of a Southern Rice Black-streaked Dwarf Virus Isolate from Maize in Northern China
2011, 26(1): 47 doi: 10.1007/s12250-011-3170-9
Received: 14 October 2010
Accepted: 06 December 2010
Southern rice black-streaked dwarf virus (SRBSDV) is a novel Fijivirus prevalent in rice in southern and central China, and northern Vietnam. Its genome has 10 segments of double-stranded RNA named S1 to S10 according to their size. An isolate of SRBSDV, JNi4, was obtained from naturally infected maize plants from Ji’ning, Shandong province, in the 2008 maize season. Segments S7 to S10 of JNi4 share nucleotide identities of 72.6%-73.1%, 72.3%-73%, 73.9%-74.5% and 77.3%-79%, respectively, with corresponding segments of Rice black-streaked dwarf virus isolates, and identities of 99.7%, 99.1%-99.7%, 98.9%-99.5%, and 98.6%-99.2% with those of SRBSDV isolates HN and GD. JNi4 forms a separate branch with GD and HN in the phylogenetic trees constructed with genomic sequences of S7 to S10. These results confirm the proposed taxonomic status of SRBSDV as a distinct species of the genus Fijivirus and indicate that JNi4 is an isolate of SRBSDV. Shandong is so far the northernmost region where SRBSDV is found in China.
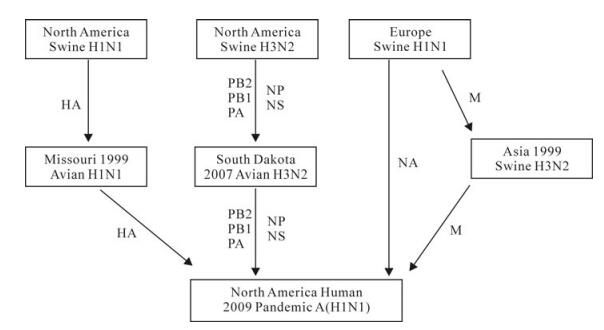
Close Relationship between the 2009 H1N1 Virus and South Dakota AIV Strains
2011, 26(1): 54 doi: 10.1007/s12250-011-3149-6
Received: 18 May 2010
Accepted: 15 October 2010
Although previous publications suggest the 2009 pandemic influenza A (H1N1) virus was reassorted from swine viruses of North America and Eurasia, the immediate ancestry still remains elusive due to the big evolutionary distance between the 2009 H1N1 virus and the previously isolated strains. Since the unveiling of the 2009 H1N1 influenza, great deal of interest has been drawn to influenza, consequently a large number of influenza virus sequences have been deposited into the public sequence databases. Blast analysis demonstrated that the recently submitted 2007 South Dakota avian influenza virus strains and other North American avian strains contained genetic segments very closely related to the 2009 H1N1 virus, which suggests these avian influenza viruses are very close relatives of the 2009 H1N1 virus. Phylogenetic analyses also indicate that the 2009 H1N1 viruses are associated with both avian and swine influenza viruses circulating in North America. Since the migrating wild birds are preferable to pigs as the carrier to spread the influenza viruses across vast distances, it is very likely that birds played an important role in the inter-continental evolution of the 2009 H1N1 virus. It is essential to understand the evolutionary route of the emerging influenza virus in order to find a way to prevent further emerging cases. This study suggests the close relationship between 2009 pandemic virus and the North America avian viruses and underscores enhanced surveillance of influenza in birds for understanding the evolution of the 2009 pandemic influenza.
Indirect ELISA with Recombinant GP5 for Detecting Antibodies to Porcine Reproductive and Respiratory Syndrome Virus
2011, 26(1): 61 doi: 10.1007/s12250-011-3154-9
Received: 22 June 2010
Accepted: 30 November 2010
Porcine reproductive and respiratory syndrome is caused by the PRRS virus (PRRSV), which has six structural proteins (GP2, GP3, GP4, GP5, M and N). GP5 and N protein are important targets for serological detection by enzyme-linked immunosorbent assay (ELISA) and other methods. Toward this goal, we developed an indirect ELISA with recombinant GP5 antigens and this method was validated by comparison to the LSI PRRSV-Ab ELISA kit. The results indicated that the optimal concentration of coated recombinant antigen was 0.2 μg/well for a serum dilution of 1:40. The rate of agreement with the LSI PRRSV-Ab kit was 88.7% (266/300). These results support the potential use of recombinant GP5 as an antigen for indirect ELISA to detect PRRSV antibodies in pigs.

Prevalence of Three Shrimp Viruses in Zhejiang Province in 2008
2011, 26(1): 67 doi: 10.1007/s12250-011-3157-6
Received: 07 July 2010
Accepted: 17 October 2010
White spot syndrome virus (WSSV), Taura syndrome virus (TSV) and Infectious hypodermal and haematopoietic necrosis virus (IHHNV) are three shrimp viruses responsible for major pandemics affecting the shrimp farming industry. Shrimps samples were collected from 12 farms in Zhejiang province, China, in 2008 and analyzed by PCR to determine the prevalence of these viruses. From the 12 sampling locations, 8 farms were positive for WSSV, 8 for IHHNV and 6 for both WSSV and IHHNV. An average percentage of 57.4% of shrimp individuals were infected with WSSV, while 49.2% were infected with IHHNV. A high prevalence of co-infection with WSSV and IHHNV among samples was detected from the following samples: Bingjiang (93.3%), liuao (66.7%), Jianshan (46.7%) and Xianxiang (46.7%). No samples exhibited evidence of infection with TSV in collected samples. This study provides comprehensive information of the prevalence of three shrimp viruses in Zhejiang and may be helpful for disease prevention control in this region
Indinavir Resistance Evolution: a Comment
2011, 26(1): 72 doi: 10.1007/s12250-011-3173-6
Received: 28 October 2010
Accepted: 16 November 2010


















