|
Hand Foot and Mouth Disease Due to Enterovirus 71 in Malaysia
2011, 26(4): 221 doi: 10.1007/s12250-011-3195-8
Received: 31 March 2011
Accepted: 24 June 2011
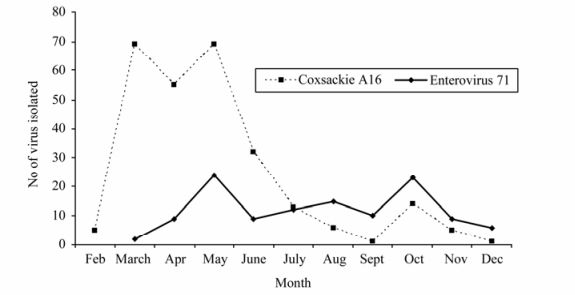
Hand foot and mouth disease is a febrile sickness complex characterized by cutaneous eruption (exanthem) on the palms and soles with simultaneous occurrence of muco-cutanous vesiculo-ulcerative lesions (enanthem) affecting the mouth. The illness is caused by a number of enteroviruses with coxsackievirus A16 and enterovirus 71 as the main causative agents. Human enterovirus 71 (EV71) belongs to the species Human enterovirus A under the genus Enterovirus within the family Picornaviridae. EV71 has been associated with an array of clinical diseases including hand foot and mouth disease (HFMD), aseptic meningitis, encephalitis and poliomyelitis-like acute flaccid paralysis. A large outbreak of HFMD due to highly neurovirulent EV71 emerged in Malaysia in 1997, and caused 41 deaths amongst young children. In late 2000, a recurrence of an outbreak of HFMD occurred in Malaysia with 8 fatalities in peninsular Malaysia. Outbreak of HFMD due to EV71 recurred in 2003 with an unknown number of cases and mortalities. A similar outbreak of HFMD with 2 recorded deaths in young children occurred in peninsular Malaysia in late 2005 and this was followed by a larger outbreak in Sarawak (Malaysian Borneo) with 6 reported fatalities in the early part of 2006. The current on-going outbreak of HFMD started in peninsular Malaysia in epidemiological week 12 of 2010. As with other HFMD outbreaks in Malaysia, both EV71 and CA16 were the main aetiological viruses isolated. In similarity with the HFMD outbreak in 2005, the isolation of CA16 preceded the appearance of EV71. Based on the VP1 gene nucleotide sequences, 4 sub-genogroups of EV71 (C1, C2, B3 and B4) co-circulated and caused the outbreak of hand, foot and mouth disease in peninsular Malaysia in 1997. Two sub-genogroups (C1 and B4) were noted to cause the outbreak in 2000 in both peninsular Malaysia and Sarawak. EV71 of sub-genogroup B5 with smaller contribution from sub-genogroup C1 caused the outbreak in 2003. In the 2005 outbreak, besides the EV71 strains of sub-genogroup C1, EV71 strains belonging to sub-genogroup B5 were isolated but formed a cluster which was distinct from the EV71 strains from the sub-genogroup B5 isolated in 2003. The four EV71 strains isolated from clinical specimens of patients with hand, foot and mouth disease in the Sarawak outbreak in early 2006 also belonged to sub-genogroup B5. Phylogenetic analysis of the VP1 gene suggests that the EV71 strains causing the outbreak in Sarawak could have originated from peninsular Malaysia. Epidemiological and molecular data since 1997 show the recurrence of HFMD due to EV71 in Malaysia every 2 to 4 years. In each of the past outbreaks, more than one sub-genogroup of the virus co-circulate.

Epidemiology and Mutational Analysis of Global Strains of Crimean-Congo Haemorrhagic Fever Virus
2011, 26(4): 229 doi: 10.1007/s12250-011-3211-z
Received: 04 July 2011
Accepted: 12 July 2011
Crimean-Congo hemorrhagic fever (CCHF) is a severe illness with high fatality. Cases are reported in several countries in Africa, Europe, the Middle East, and Asia. Phylogenetic analyses based on the virus S (nucleocapsid), M (glycoprotein), and L (polymerase) genome segments sequences indicate distinct geographic lineages exist but their specific genetic characteristics require elucidation. In this work we collected all full length S segment sequences and generated a phylogenetic tree based on the alignment of these 62 samples. We then analyzed the alignment using entries from AAIndex, the Amino Acid Index database, to identify amino acid mutations that performed significant changes in charge, pka, hydropathy and side chain volume. Finally, we mapped these changes back to the tree and alignment to identify correlated mutations or sites that characterized a specific lineage. Based on this analysis we are able to propose a number of sites that appear to be important for virus function and which would be good candidates for experimental mutational analysis studies.
Rapid and Sensitive Detection of PRRSV by a Reverse Transcription-Loop-mediated Isothermal Amplification Assay
2011, 26(4): 252 doi: 10.1007/s12250-011-3185-x
Received: 24 January 2011
Accepted: 11 May 2011
A real-time monitoring reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay was developed for the sensitive and specific detection of prototypic, prevalent North American porcine reproductive and respiratory syndrome virus (PRRSV) strains. As a higher sensitivity and specificity method than reverse transcription polymerase chain reaction (RT-PCR), the RT-LAMP method only used a turbidimeter, exhibited a detection limit corresponding to a 10–4 dilution of template RNA extracted from 250 μL of 105 of the 50% tissue culture infective dose (TCID50) of PRRSV-containing cells, and no cross-reactivity was observed with other related viruses including porcine circovirus type 2, swine influenza virus, porcine rotavirus and classical swine fever virus. From forty-two field samples, 33 samples in the RT-LAMP assay was detected positive, whereas three of which were not detected by RT-PCR. Furthermore, in 33 strains of PRRSV, an identical detection rate was observed with the RT-LAMP assay to what were isolated using porcine alveolar macrophages.These findings demonstrated that the RT-LAMP assay has potential clinical applications for the detection of highly pathogenic PRRSV isolates, especially in developing countries.
Characterization and Diagnostic Use of a Monoclonal Antibody for VP28 Envelope Protein of White Spot Syndrome Virus *
2011, 26(4): 260 doi: 10.1007/s12250-011-3202-0
Received: 17 May 2011
Accepted: 29 June 2011
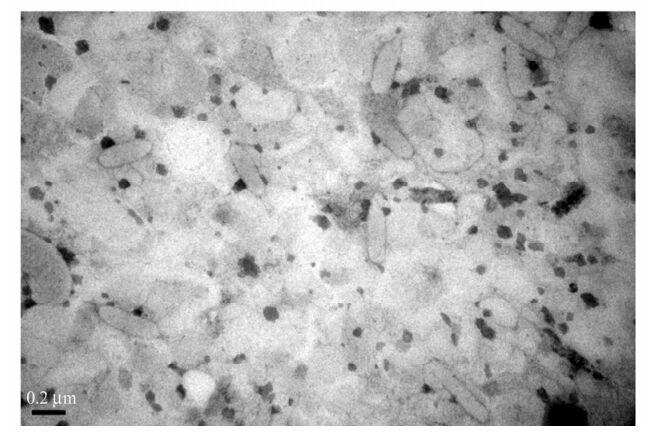
The gene encoding the VP28 envelope protein of White spot syndrome virus (WSSV) was cloned into expression vector pET-30a and transformed into the Escherichia coli strain BL21. After induction, the recombinant VP28 (rVP28) protein was purified and then used to immunize Balb/c mice for monoclonal antibody (MAb) production. It was observed by immuno-electron microscopy the MAbs specific to rVP28 could recognize native VP28 target epitopes of WSSV and dot-blot analysis was used to detect natural WSSV infection. Competitive PCR showed that the viral level was approximately 104 copies/mg tissue in the dilution of gill homogenate of WSSV-infected crayfish at the detection limit of dot-blot assay. Our results suggest that dot-blot analysis with anti-rVP28 MAb could rapidly and sensitively detect WSSV at the early stages of WSSV infection.

Application of GP5 Protein to Develop Monoclonal Antibody against Porcine Reproductive and Respiratory Syndrome Virus *
2011, 26(4): 267 doi: 10.1007/s12250-011-3198-5
Received: 27 April 2011
Accepted: 27 June 2011
In this study, a panel of monoclonal antibodies (mAbs) against Porcine reproductive and respiratory syndrome virus(PRRSV), named as 8C9 and4B4, were produced by fusing SP2/0 myeloma cells and spleen cells of BALB/c mice immunized with the PRRSV (TCID50=5.5), screened by the indirect ELISA and subjected to several limiting dilutions. mAbs were then identified by biological characterization. Among the two fusion cell strains, 8C9 belonged to the IgG1 subclass and 4B4 belonged to the IgG2a subclass. The titers in cell culture supernatant and abdomen liquor reached to 1:104and 1:105, respectively. The specificity test indicated that the two cells had specific reactions for the PRRSV and GP5 protein respectively, and no reaction with Classical swine fever virus (CSFV) or Swine vesicular disease virus (SVDV). The molecular weights of the heavy chain and light chain were about 45.0 kDa and 25.0 kDa, respectively. In neutralization activity tests, the results showed that the prepared mAb 4B4 can protect 50% of cells with no CPE in dilution up to 1:512, but mAB 8C9 has no neutralization activities to PRRSV.
Develope Monoclonal Antibody against Foot-and-mouth Disease Virus A Type *
2011, 26(4): 273 doi: 10.1007/s12250-011-3199-4
Received: 04 May 2011
Accepted: 07 July 2011
In order to develop an anti-FMDV A Type monoclonal antibody (mAb), BABL/c mice were immunized with FMDV A type. Monoclonal antibodies (mAbs) 7B11 and 8H4 against Foot-and-mouth disease virus (FMDV) serotype A were produced by fusing SP2/0 myeloma cells with splenocyte from the mouse immunized with A/AV88. The microneutralization titer of the mAbs 7B11 and 8H4 were 1024 and 512, respectively. Both mAbs contain kappa light chains, the mAbs were IgG1. In order to define the mAbs binding epitopes, the reactivity of these mAbs against A Type FMDV, were examined using indirect ELISA, the result showed that both mAbs reacted with A Type FMDV. These mAbs may be used for further vaccine studies, diagnostic methods, prophylaxis, etiological and immunological research on FMDV. Characterization of these ncindicated that prepared ant i-FMDV A mAbs had no cross-reactivity with Swine Vesicular Disease (SVD) or FMDV O, Asia1 and C Type antigens. Their titers in abdomen liquor were 1:5×106 and 1:2×106, respectively. 7B11 was found to be of subtype IgG1, 8H4 was classified as IgG2b subtype. The mAbs prepared in this study, are specific for detection of FMDV serotype A, and is potentially useful for pen-side diagnosis.
Bioinformatic Analysis of BBTV Satellite DNA in Hainan
2011, 26(4): 279 doi: 10.1007/s12250-011-3196-7
Received: 09 April 2011
Accepted: 01 July 2011
Banana bunchy top virus (BBTV), family Nanaviridae, genus Babuvirus, is a single stranded DNA virus (ssDNA) that causes banana bunchy top disease (BBTD) in banana plants. It is the most common and most destructive of all viruses in these plants and is widespread throughout the Asia-Pacific region. In this study we isolated, cloned and sequenced a BBTV sample from Hainan Island, China. The results from sequencing and bioinformatics analysis indicate this isolate represents a satellite DNA component with 12 DNA sequences motifs. We also predicted the physical and chemical properties, structure, signal peptide, phosphorylation, secondary structure, tertiary structure and functional domains of its encoding protein, and compare them with the corresponding quantities in the replication initiation protein of BBTV DNA1.

Hemorrhagic Fever with Renal Syndrome Complicated by Orchitis
2011, 26(4): 285 doi: 10.1007/s12250-011-3182-0
Received: 12 January 2011
Accepted: 13 May 2011
Hemorrhagic fever with renal syndrome (HFRS) is a disease caused by viruses of the family Bunyaviridae, genus Hantavirus. HFRS from Dobrava virus (DOBV) is a seldom reported disease in Albania. Clinically HFRS is manifested as mild, moderate, or severe. Therefore, the number of cases of Hantavirus’ infection may be underestimated, and should be included in the differential diagnosis of many acute infections, hematologic diseases, acute abdominal diseases and renal diseases complicated by acute renal failure. We report here an atypical presentation of HFRS from Dobrava virus complicated by orchitis with a positive outcome.
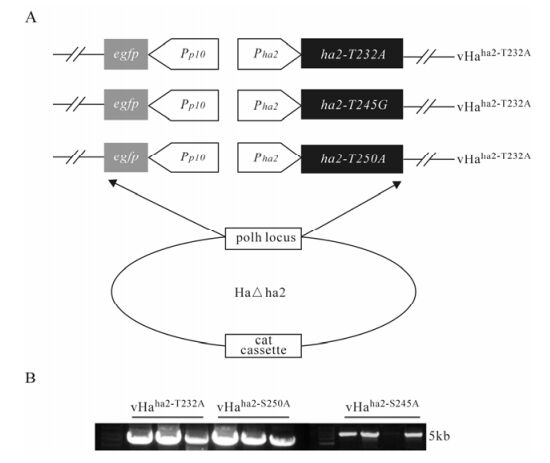
Putative Phosphorylation Sites On WCA Domain of HA2 Is Essential For Helicoverpa armigera Single Nucleopolyhedrovirus Replication *
2011, 26(4): 245 doi: 10.1007/s12250-011-3189-6
Received: 09 February 2011
Accepted: 25 February 2011
Protein phosphorylation is one of the most common post-translational modification processes that play an essential role in regulating protein functionality. The Helicoverpa armigera single nucleopolyhedrovirus (HearNPV) orf2-encoded nucleocapsid protein HA2 participates in orchestration of virus-induced actin polymerization through its WCA domain, in which phosphorylation status are supposed to be critical in respect to actin polymerization. In the present study, two putative phosphorylation sites (232Thr and 250Ser) and a highly conserved Serine (245Ser) on the WCA domain of HA2 were mutated, and their phenotypes were characterized by reintroducing the mutated HA2 into the HearNPV genome. Viral infectivity assays demonstrated that only the recombinant HearNPV bearing HA2 mutation at 245Ser can produce infectious virions, both 232Thr and 250Ser mutations were lethal to the virus. However, actin polymerization assay demonstrated that all the three viruses bearing HA2 mutations were still capable of initiating actin polymerization in the host nucleus, which indicated the putative phosphorylation sites on HA2 may contribute to HearNPV replication through another unidentified pathway.


















