
2021 Vol.36(2)
African swine fever (ASF), a highly lethal swine disease caused by African swine fever virus (ASFV), has spread to many countries of Africa, Europe and Asia, posing a huge threat to the global swine industry and food security. But no commercial vaccines or antivirals are currently available against ASFV. We are facing the task to further explore the biology of the virus and develop effective control measurements. This issue presents a collection of interesting research progress on ASFV, which reveals viral protein functions and cellular interactions, viral epigenome features, vaccine strategies, as well as new diagnostic methods. We hope those findings could increase our knowledge and provide valuable reference for future research of ASFV interventions.
|
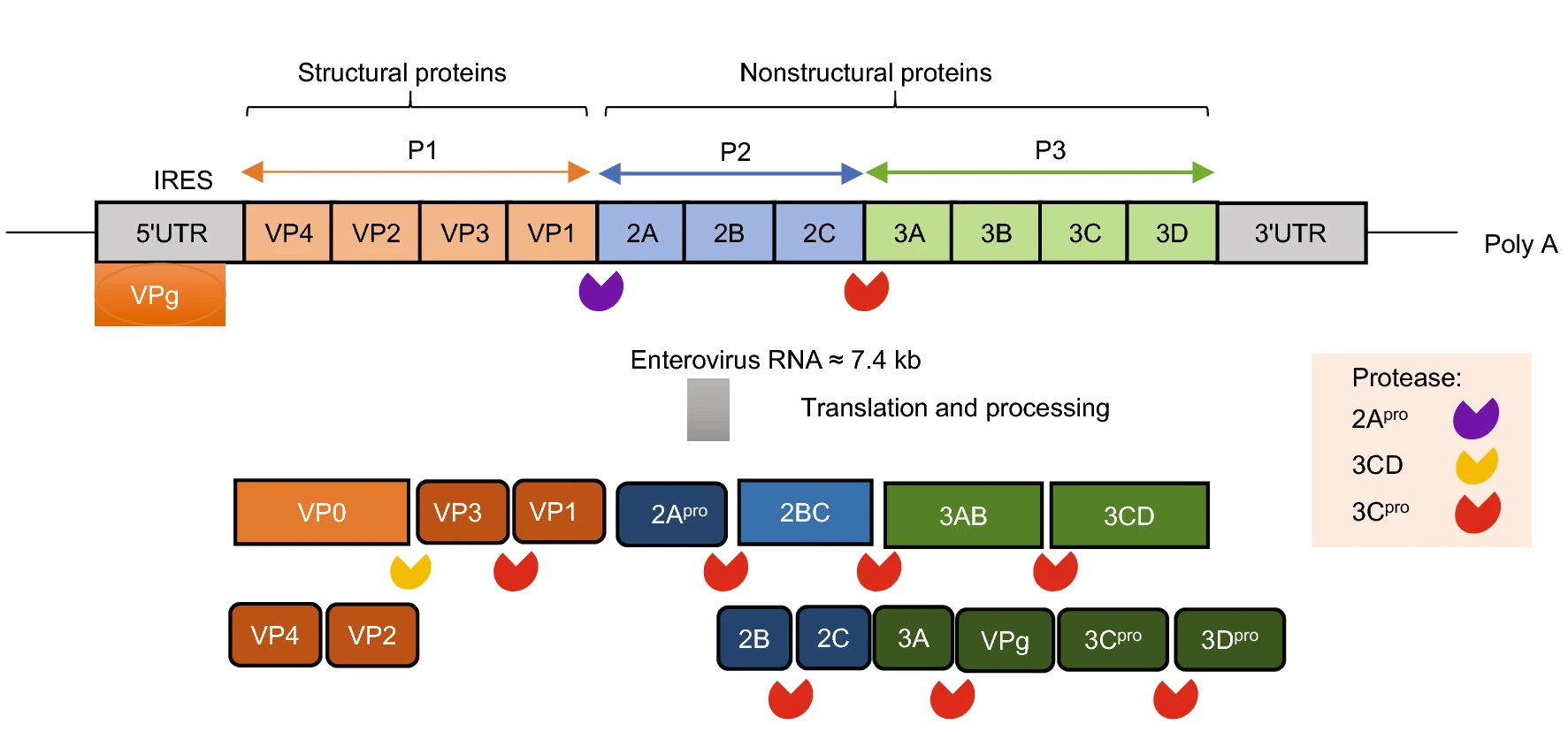
From Monovalent to Multivalent Vaccines, the Exploration for Potential Preventive Strategies Against Hand, Foot, and Mouth Disease (HFMD)
2021, 36(2): 167 doi: 10.1007/s12250-020-00294-3
Hand, foot, and mouth disease (HFMD) recently emerged as a global public threat. The licensure of inactivated enterovirus A71 (EV-A71) vaccine was the first step in using a vaccine to control HFMD. New challenges arise from changes in the pathogen spectrum while vaccines directed against other common serotypes are in the preclinical stage. The mission of a broad-spectrum prevention strategy clearly favors multivalent vaccines. The development of multivalent vaccines was attempted via the simple combination of potent monovalent vaccines or the construction of chimeric vaccines comprised of epitopes derived from different virus serotypes. The present review summarizes recent advances in HFMD vaccine development and discusses the next steps toward a safe and effective HFMD vaccine that is capable of establishing a cross-protective antibody response.

African Swine Fever Virus MGF360-12L Inhibits Type I Interferon Production by Blocking the Interaction of Importin α and NF-кB Signaling Pathway
2021, 36(2): 176 doi: 10.1007/s12250-020-00304-4
African swine fever (ASF) is an infectious transboundary disease of domestic pigs and wild boar and spreading throughout Eurasia. There is no vaccine and treatment available. Complex immune escape strategies of African swine fever virus (ASFV) are crucial factors affecting immune prevention and vaccine development. MGF360 genes have been implicated in the modulation of the IFN-I response. The molecular mechanisms contributing to innate immunity are poorly understood. In this study, we demonstrated that ASFV MGF360-12L (MGF360 families 12L protein) significantly inhibited the mRNA transcription and promoter activity of IFN-β and NF-кB, accompanied by decreases of IRF3, STING, TBK1, ISG54, ISG56 and AP-1 mRNA transcription. Also, MGF360-12L might suppress the nuclear localization of p50 and p65 mediated by classical nuclear localization signal (NLS). Additionally, MGF360-12L could interact with KPNA2, KPNA3, and KPNA4, which interrupted the interaction between p65 and KPNA2, KPNA3, KPNA4. We further found that MGF360-12L could interfere with the NF-кB nuclear translocation by competitively inhibiting the interaction between NF-кB and nuclear transport proteins. These findings suggested that MGF360-12L could inhibit the IFN-I production by blocking the interaction of importin a and NF-кB signaling pathway, which might reveal a novel strategy for ASFV to escape the host innate immune response.
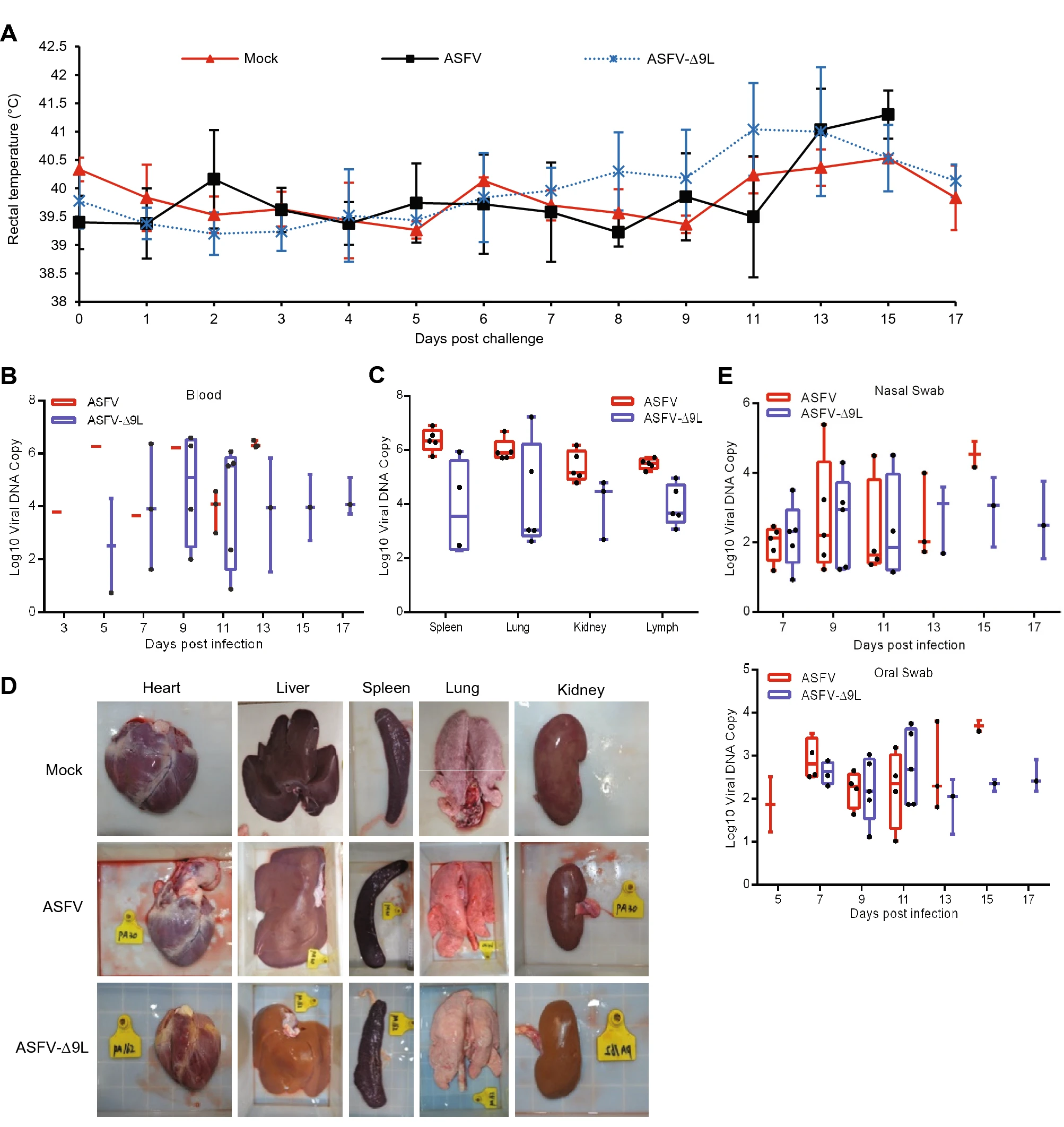
African Swine Fever Virus MGF-110-9L-deficient Mutant Has Attenuated Virulence in Pigs
2021, 36(2): 187 doi: 10.1007/s12250-021-00350-6
African swine fever virus (ASFV) is the etiological agent of African swine fever (ASF), an often lethal disease in domestic and wild pigs. ASF represents a major threat to the swine industry worldwide. Currently, no commercial vaccine is available because of the complexity of ASFV or biosecurity concerns. Live attenuated viruses that are naturally isolated or genetically manipulated have demonstrated reliable protection against homologous ASFV strain challenge. In the present study, a mutant ASFV strain with the deletion of ASFV MGF-110-9L (ASFV-Δ9L) was generated from a highly virulent ASFV CN/GS/2018 parental strain, a genotype II ASFV. Relative to the parental ASFV isolate, deletion of the MGF-110-9L gene significantly decreased the ability of ASFV-Δ9L to replicate in vitro in primary swine macrophage cell cultures. The majority of animals inoculated intramuscularly with a low dose of ASFV-Δ9L (10 HAD50) remained clinically normal during the 21-day observational period. Three of five ASFV-Δ9L-infected animals displayed low viremia titers and low virus shedding and developed a strong virus-specific antibody response, indicating partial attenuation of the ASFV-Δ9L strain in pigs. The findings imply the potential usefulness of the ASFV-Δ9L strain for further development of ASF control measures.
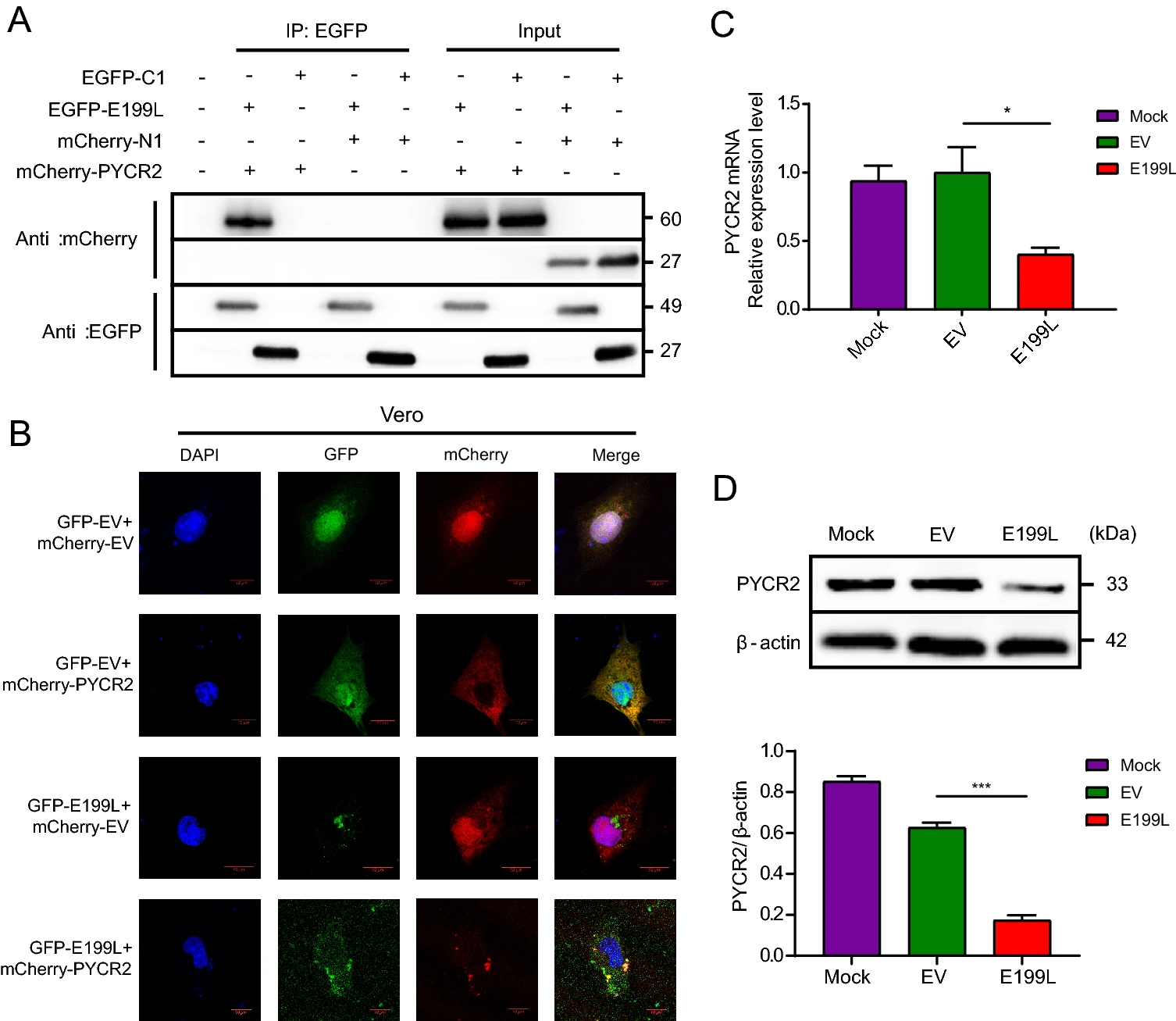
African Swine Fever Virus Protein E199L Promotes Cell Autophagy through the Interaction of PYCR2
2021, 36(2): 196 doi: 10.1007/s12250-021-00375-x
African swine fever virus (ASFV), as a member of the large DNA viruses, may regulate autophagy and apoptosis by inhibiting programmed cell death. However, the function of ASFV proteins has not been fully elucidated, especially the role of autophagy in ASFV infection. One of three Pyrroline-5-carboxylate reductases (PYCR), is primarily involved in conversion of glutamate to proline. Previous studies have shown that depletion of PYCR2 was related to the induction of autophagy. In the present study, we found for the first time that ASFV E199L protein induced a complete autophagy process in Vero and HEK-293T cells. Through co-immunoprecipitation coupled with mass spectrometry (CoIP-MS) analysis, we firstly identified that E199L interact with PYCR2 in vitro. Importantly, our work provides evidence that E199L down-regulated the expression of PYCR2, resulting in autophagy activation. Overall, our results demonstrate that ASFV E199L protein induces complete autophagy through interaction with PYCR2 and down-regulate the expression level of PYCR2, which provide a valuable reference for the role of autophagy during ASFV infection and contribute to the functional clues of PYCR2.

Porcine Immunoglobulin Fc Fused P30/P54 Protein of African Swine Fever Virus Displaying on Surface of S. cerevisiae Elicit Strong Antibody Production in Swine
2021, 36(2): 207 doi: 10.1007/s12250-020-00278-3
African swine fever virus (ASFV) infects domestic pigs and European wild boars with strong, hemorrhagic and high mortality. The primary cellular targets of ASFV is the porcine macrophages. Up to now, no commercial vaccine or effective treatment available to control the disease. In this study, three recombinant Saccharomyces cerevisiae (S. cerevisiae) strains expressing fused ASFV proteins-porcine Ig heavy chains were constructed and the immunogenicity of the S. cerevisiae-vectored cocktail ASFV feeding vaccine was further evaluated. To be specific, the P30-Fcγ and P54-Fcα fusion proteins displaying on surface of S. cerevisiae cells were produced by fusing the Fc fragment of porcine immunoglobulin IgG1 or IgA1 with p30 or p54 gene of ASFV respectively. The recombinant P30-Fcγ and P54-Fcα fusion proteins expressed by S. cerevisiae were verified by Western blotting, flow cytometry and immunofluorescence assay. Porcine immunoglobulin Fc fragment fused P30/P54 proteins elicited P30/P54-specific antibody production and induced higher mucosal immunity in swine. The absorption and phagocytosis of recombinant S. cerevisiae strains in IPEC-J2 cells or porcine alveolar macrophage (PAM) cells were significantly enhanced, too. Here, we introduce a kind of cheap and safe oral S. cerevisiae-vectored vaccine, which could activate the specific mucosal immunity for controlling ASFV infection.
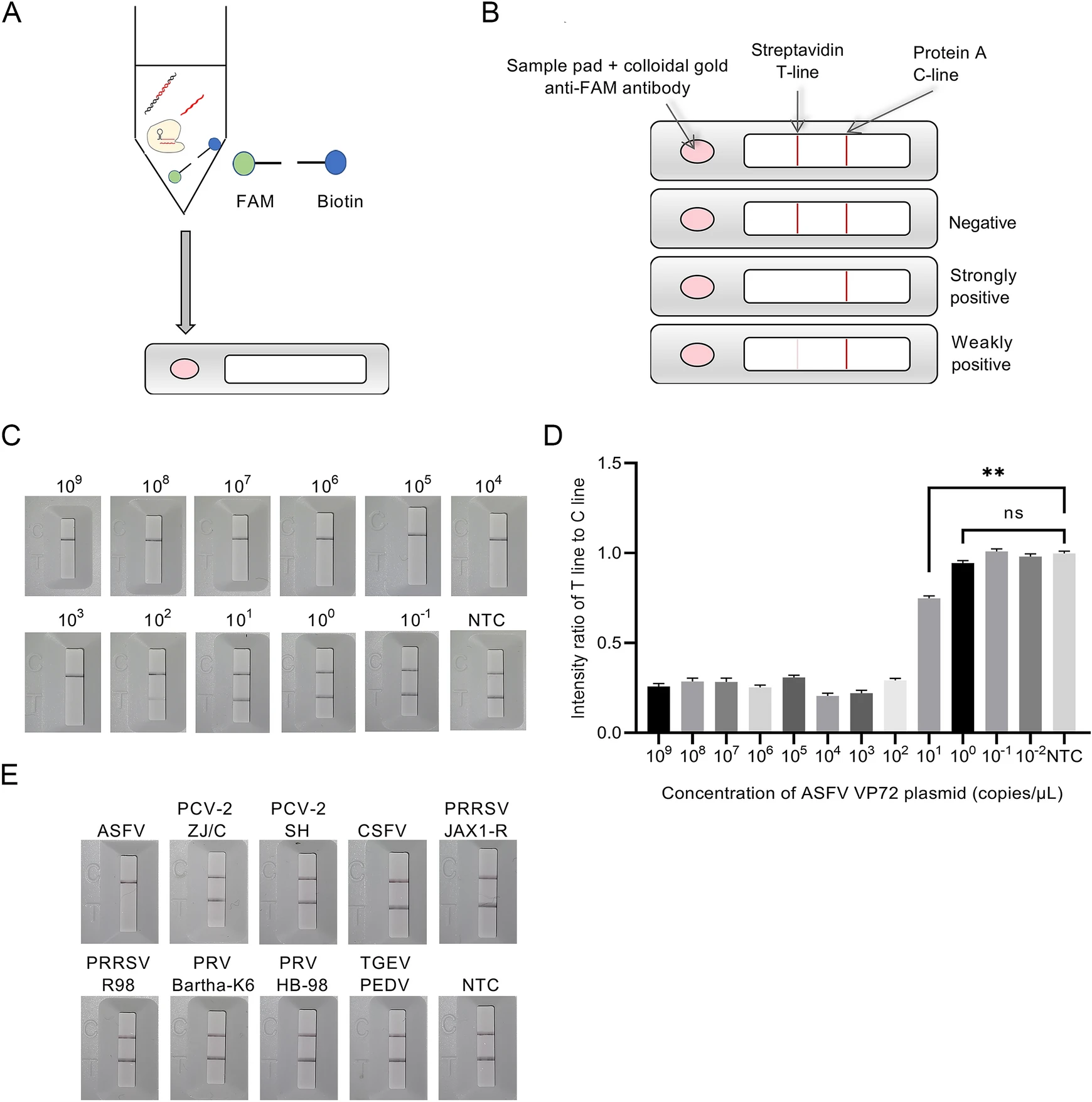
Development of A Super-Sensitive Diagnostic Method for African Swine Fever Using CRISPR Techniques
2021, 36(2): 220 doi: 10.1007/s12250-020-00323-1
African swine fever (ASF) is an infectious disease caused by African swine fever virus (ASFV) with clinical symptoms of high fever, hemorrhages and high mortality rate, posing a threat to the global swine industry and food security. Quarantine and control of ASFV is crucial for preventing swine industry from ASFV infection. In this study, a recombinase polymerase amplification (RPA)-CRISPR-based nucleic acid detection method was developed for diagnosing ASF. As a highly sensitive method, RPA-CRISPR can detect even a single copy of ASFV plasmid and genomic DNA by determining fluorescence signal induced by collateral cleavage of CRISPR-lwCas13a (previously known as C2c2) through quantitative real-time PCR (qPCR) and has the same or even higher sensitivity than the traditional qPCR method. A lateral flow strip was developed and used in combination with RPA-CRISPR for ASFV detection with the same level of sensitivity of TaqMan qPCR. Likewise, RPA-CRISPR is capable of distinguishing ASFV genomic DNA from viral DNA/RNA of other porcine viruses without any cross-reactivity. This diagnostic method is also available for diagnosing ASFV clinical DNA samples with coincidence rate of 100% for both ASFV positive and negative samples. RPA-CRISPR has great potential for clinical quarantine of ASFV in swine industry and food security.
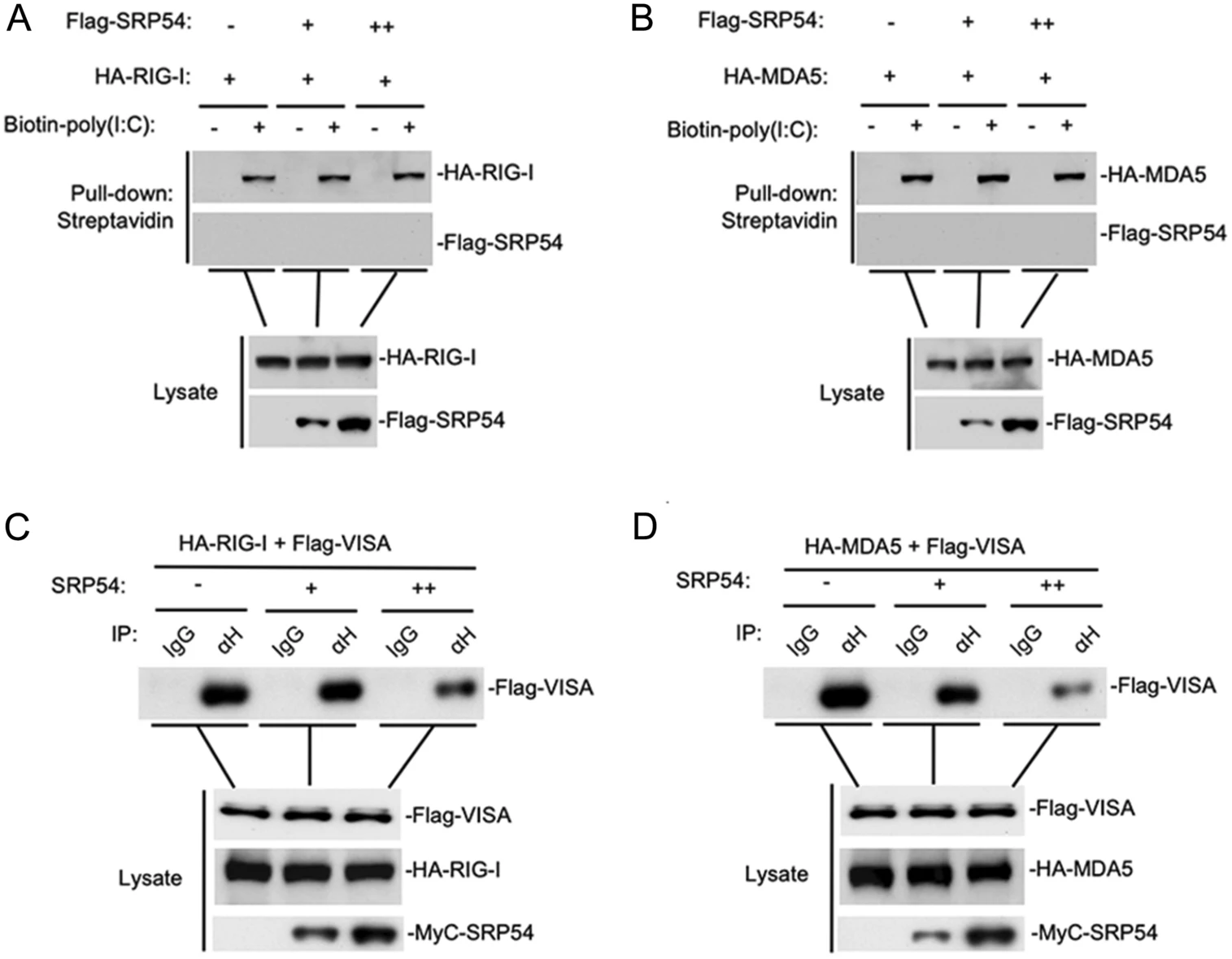
SRP54 Negatively Regulates IFN-Beta Production and Antiviral Response by Targeting RIG-I and MDA5
2021, 36(2): 231 doi: 10.1007/s12250-020-00267-6
During virus infection, RIG-I-like receptors (RLRs) recognize viral RNAs and recruit the adaptor protein VISA to activate downstream signaling, leading to activation of transcription factors NF-κB and IRF3, which collaborate to induce type I interferons (IFNs). IFNs further induce expression of hundreds of IFN-stimulated genes (ISGs) that suppress viral replication and facilitate the adaptive immune response. Dysregulated production of IFNs is implicated in various immune diseases. Here we identified Signal Recognition Particle 54 (SRP54) as a negative regulator of RLRs-induced antiviral signaling. Overexpression of SRP54 inhibited RNA virus-triggered induction of IFN-β and increased viral replication, whereas knockdown of SRP54 had opposite effects. Mechanistically, SRP54 interacted with both RIG-I and MDA5 and impaired their association with VISA. Our findings demonstrate that SRP54 acts as a negative regulator of RLRs-mediated innate immune response by disrupting the recruitment of VISA to RIG-I/MDA5.
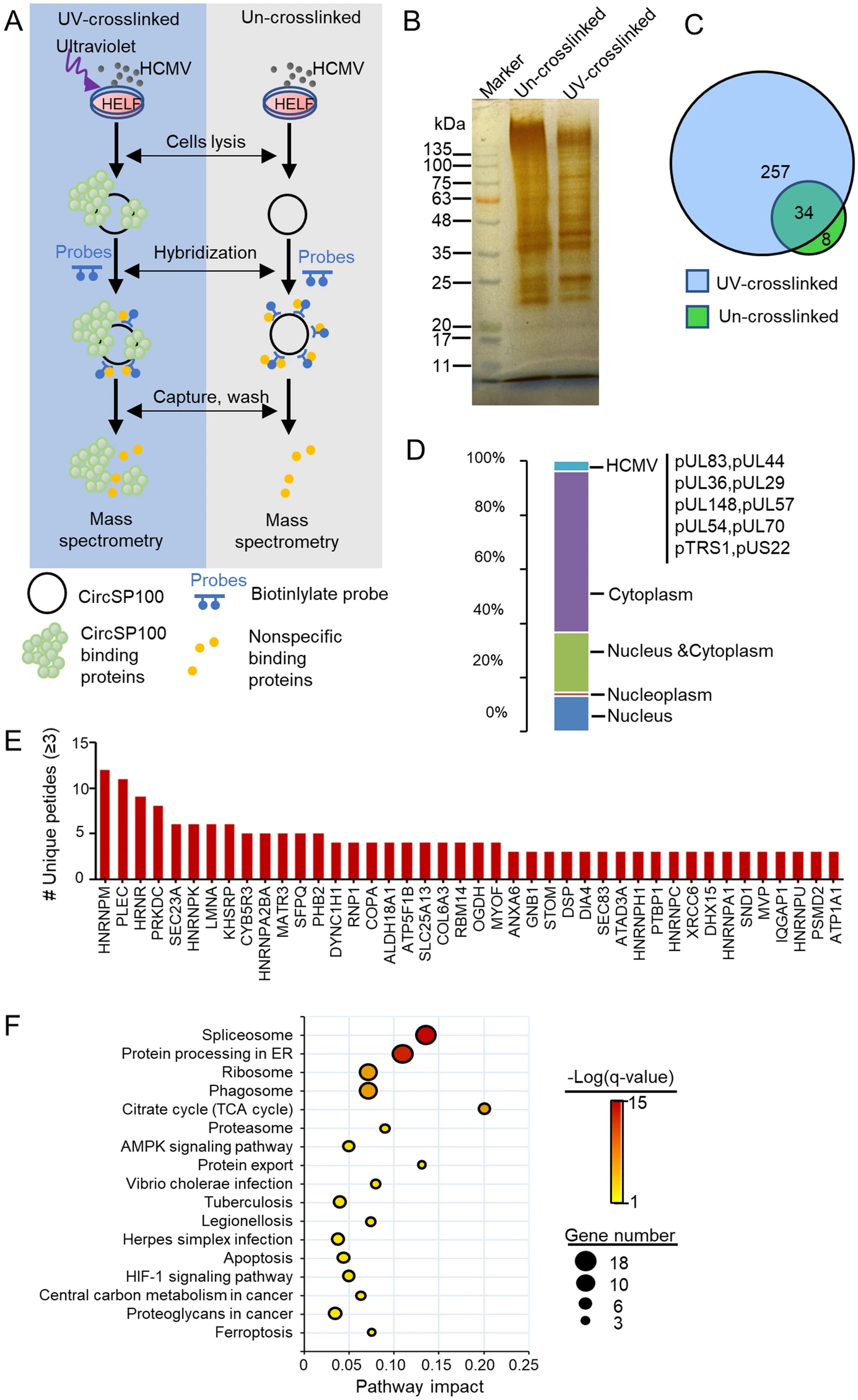
Human Cytomegalovirus Influences Host circRNA Transcriptions during Productive Infection
2021, 36(2): 241 doi: 10.1007/s12250-020-00275-6
Human cytomegalovirus (HCMV) is a double-strand DNA virus widely infected in human. Circular RNAs (circRNAs) are non-coding RNAs with most functions of which keep unknown, and the effects of HCMV productive infection on host circRNA transcriptions remain unclear. In this study, we profiled 283 host circRNAs that significantly altered by HCMV productive infection in human embryonic lung fibroblasts (HELF) by RNA deep sequencing and bioinformatics analysis. Among these, circSP100, circMAP3K1, circPLEKHM1, and circTRIO were validated for their transcriptions and sequences. Furthermore, characteristics of circSP100 were investigated by RT-qPCR and northern blot. It was implied that circSP100 was produced from the sense strand of the SP100 gene containing six exons. Kinetics of circSP100 and SP100 mRNA were significantly different after infection: circSP100 levels increased gradually along with infection, whereas SP100 mRNA levels increased in the beginning and dropped at 24 h post-infection (hpi). Meanwhile, a total number of 257 proteins, including 10 HCMV encoding proteins, were identified potentially binding to cytoplasmic circSP100 by RNA antisense purification (RAP) and mass spectrometry. Enrichment analysis showed these proteins were mainly involved in the spliceosome, protein processing, ribosome, and phagosome pathways, suggesting multiple functions of circSP100 during HCMV infection.
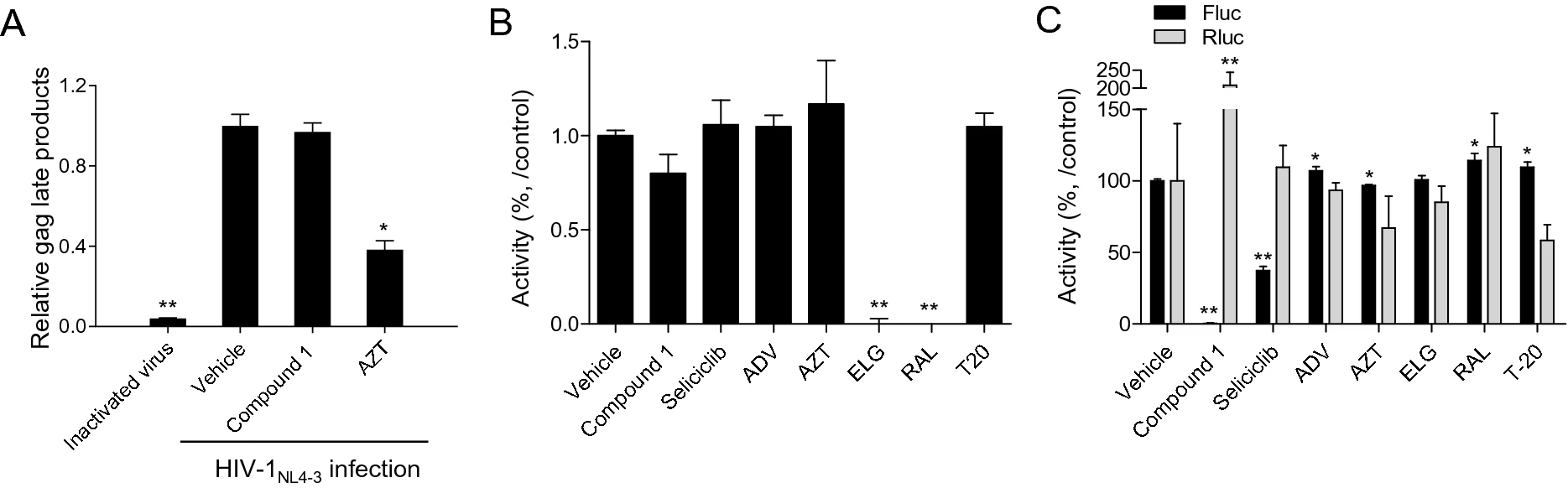
Identification of Aristolactam Derivatives That Act as Inhibitors of Human Immunodeficiency Virus Type 1 Infection and Replication by Targeting Tat-Mediated Viral Transcription
2021, 36(2): 254 doi: 10.1007/s12250-020-00274-7
Despite the success of antiretroviral therapy (ART), efforts to develop new classes of antiviral agents have been hampered by the emergence of drug resistance. Dibenzo-indole-bearing aristolactams are compounds that have been isolated from various plants species and which show several clinically relevant effects, including anti-inflammatory, antiplatelet, and anti-mycobacterial actions. However, the effect of these compounds on human immunodeficiency virus type 1 (HIV-1) infection has not yet been studied. In this study, we discovered an aristolactam derivative bearing dibenzo[cd, f]indol-4(5H)-one that had a potent anti-HIV-1 effect. A structure-activity relationship (SAR) study using nine synthetic derivatives of aristolactam identified the differing effects of residue substitutions on the inhibition of HIV-1 infection and cell viability. Among the compounds tested,1,2,8,9-tetramethoxy-5-(2-(piperidin-1-yl)ethyl)-dibenzo[cd, f]indol-4(5H)-one (Compound 2) exhibited the most potent activity by inhibiting HIV-1 infection with a half-maximal inhibitory concentration (IC50) of 1.03 μmol/L and a half-maximal cytotoxic concentration (CC50) of 16.91 μmol/L (selectivity index,16.45). The inhibitory effect of the compounds on HIV-1 infection was linked to inhibition of the viral replication cycle. Mode-of-action studies showed that the aristolactam derivatives did not affect reverse transcription or integration; instead, they specifically inhibited Tat-mediated viral transcription. Taken together, these findings show that several aristolactam derivatives impaired HIV-1 infection by inhibiting the activity of Tat-mediated viral transcription, and suggest that these derivatives could be antiviral drug candidates.
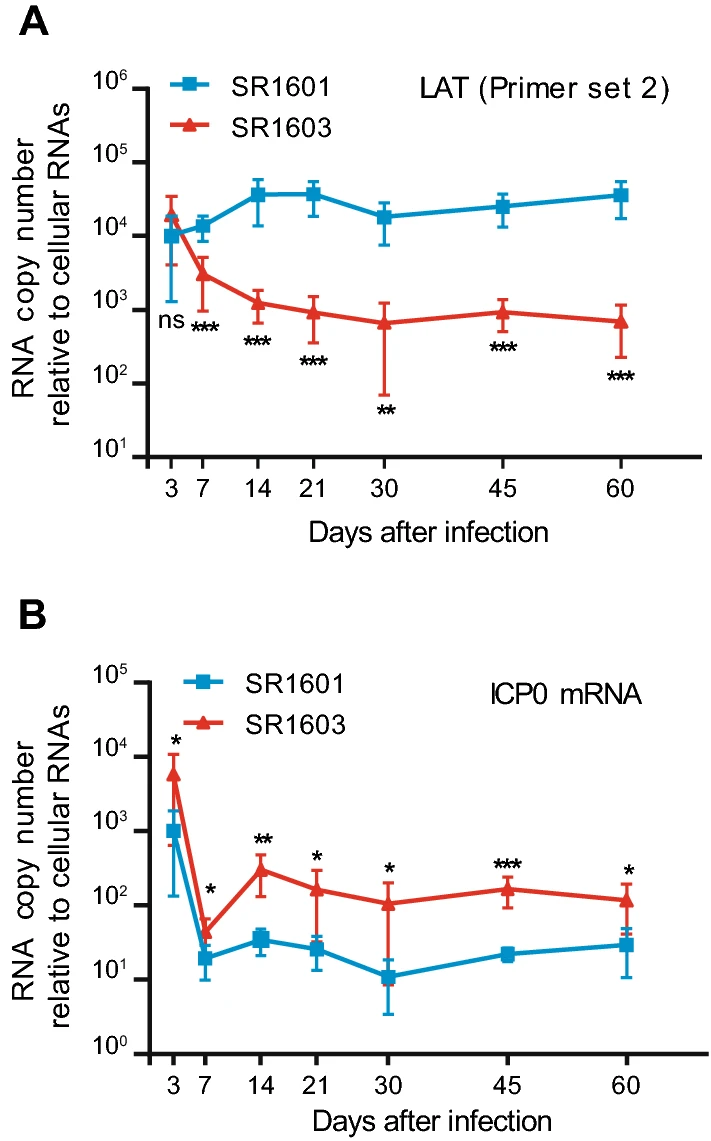
Termination of Transcription of LAT Increases the Amounts of ICP0 mRNA but Does Not Alter the Course of HSV-1 Infection in Latently Infected Murine Ganglia
2021, 36(2): 264 doi: 10.1007/s12250-020-00287-2
On entering sensory ganglia, herpes simplex viruses 1 (HSV-1) establishes a latent infection with the synthesis of a latency associated transcript (LAT) or initiates productive infection with expression of a set of immediate early viral proteins. The precise mechanisms how expression of α genes is suppressed during the latency are unknown. One mechanism that has been proposed is illustrated in the case of ICP0, a key immediate early viral regulatory protein. Specifically, the 2 kb LAT intron is complementary to the 3′ terminal portion of ICP0 mRNA. To test the hypothesis that accumulation of LAT negatively affects the accumulation of ICP0 mRNA, we inserted a DNA fragment encoding two poly(A) sequences into LAT to early terminate LAT transcript without interrupting the complementary sequence of ICP0 transcript (named as SR 1603). Comparisons of the parent (SR 1601) and mutant (SR 1603) HSV-1 viruses showed the following: Neurons harboring latent SR1603 virus accumulated equivalent amounts of viral DNA but higher amounts of ICP0 mRNA and lower amounts of LAT, when compared to neurons harboring the SR1601 virus. One notable difference between the two viruses is that viral RNA accumulation in explanted ganglia harboring SR1603 virus initiated significantly sooner than that in neurons harboring SR1601 virus, suggesting that ICP0 may act as an activator of viral gene expression in permissive cells. Collectively, these data suggest that increased ICP0 mRNA by suppressed LAT did not affect the establishment of latency in latently infected murine ganglia.
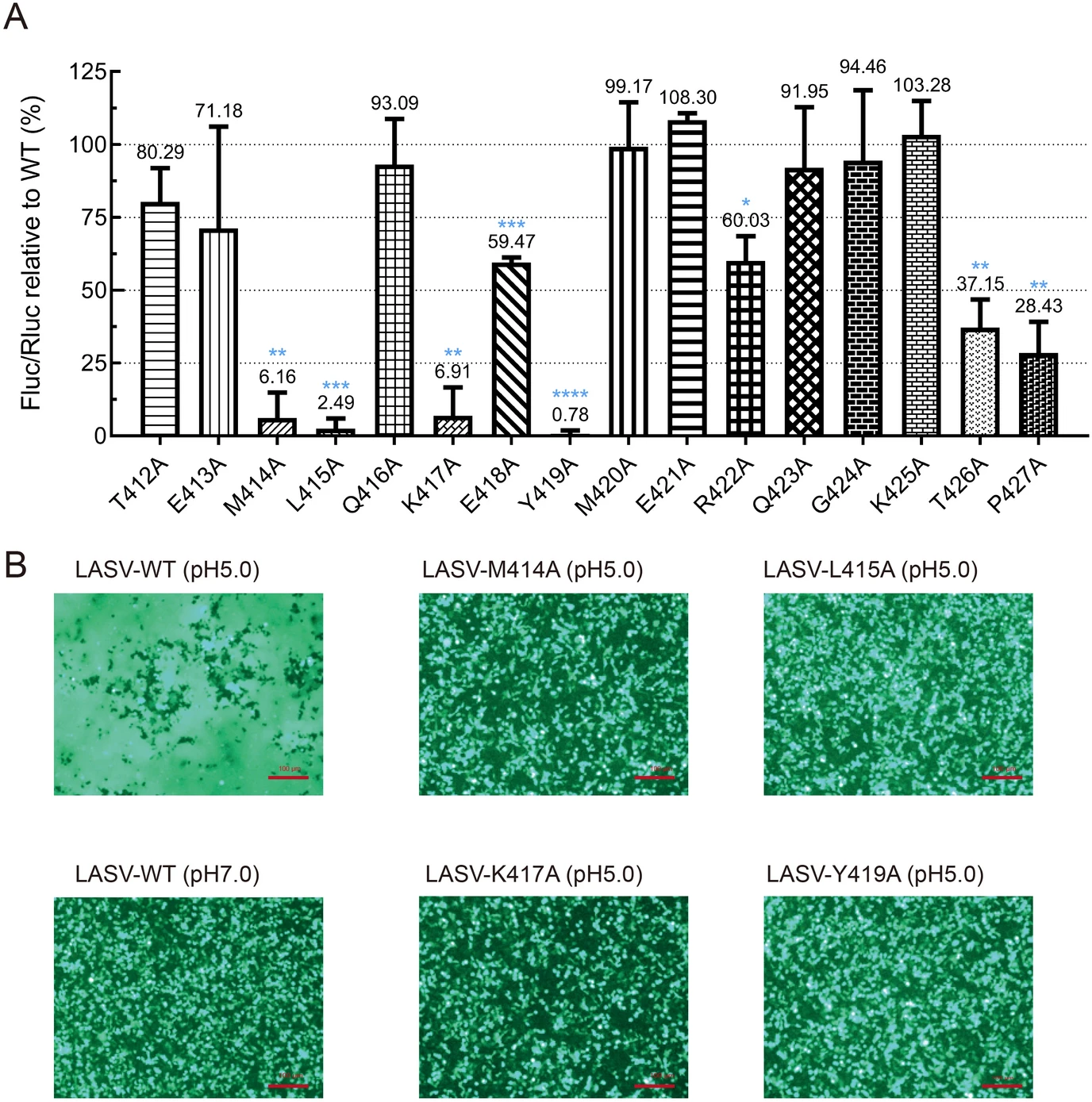
Characterizing the Lassa Virus Envelope Glycoprotein Membrane Proximal External Region for Its Role in Fusogenicity
2021, 36(2): 273 doi: 10.1007/s12250-020-00286-3
The membrane-proximal external region (MPER) of Lassa virus (LASV) glycoprotein complex (GPC) is critical in modulating its functionality. Till now, the high-resolution structure of the intact GPC, including MPER is not available. In this study, we used alanine substitution to scan all 16 residues located in LASV MPER. Western blotting and quantification fusion assay showed that the residues located at the C terminus of the HR2 (M414 and L415) and N terminus of the MPER (K417 and Y419) are critical for GPC-mediated membrane fusion function. Furthermore, cell surface biotinylation experiments revealed that M414A, K417A and Y419A expressed similar levels as WT, whereas L415A mutant led to a reduction of mature GPC on the cell surface. Moreover, substitution of these residues with the similar residue such as M414L, L415I, K417R and Y419F would partly compensate the loss of the fusion activity caused by the alanine mutant in these sites. Results from this study showed that several key residues in the MPER region are indispensable to promote the conformational changes that drive fusion events and shed light on the structure analysis of LASV GPC and anti-LASV therapeutics.
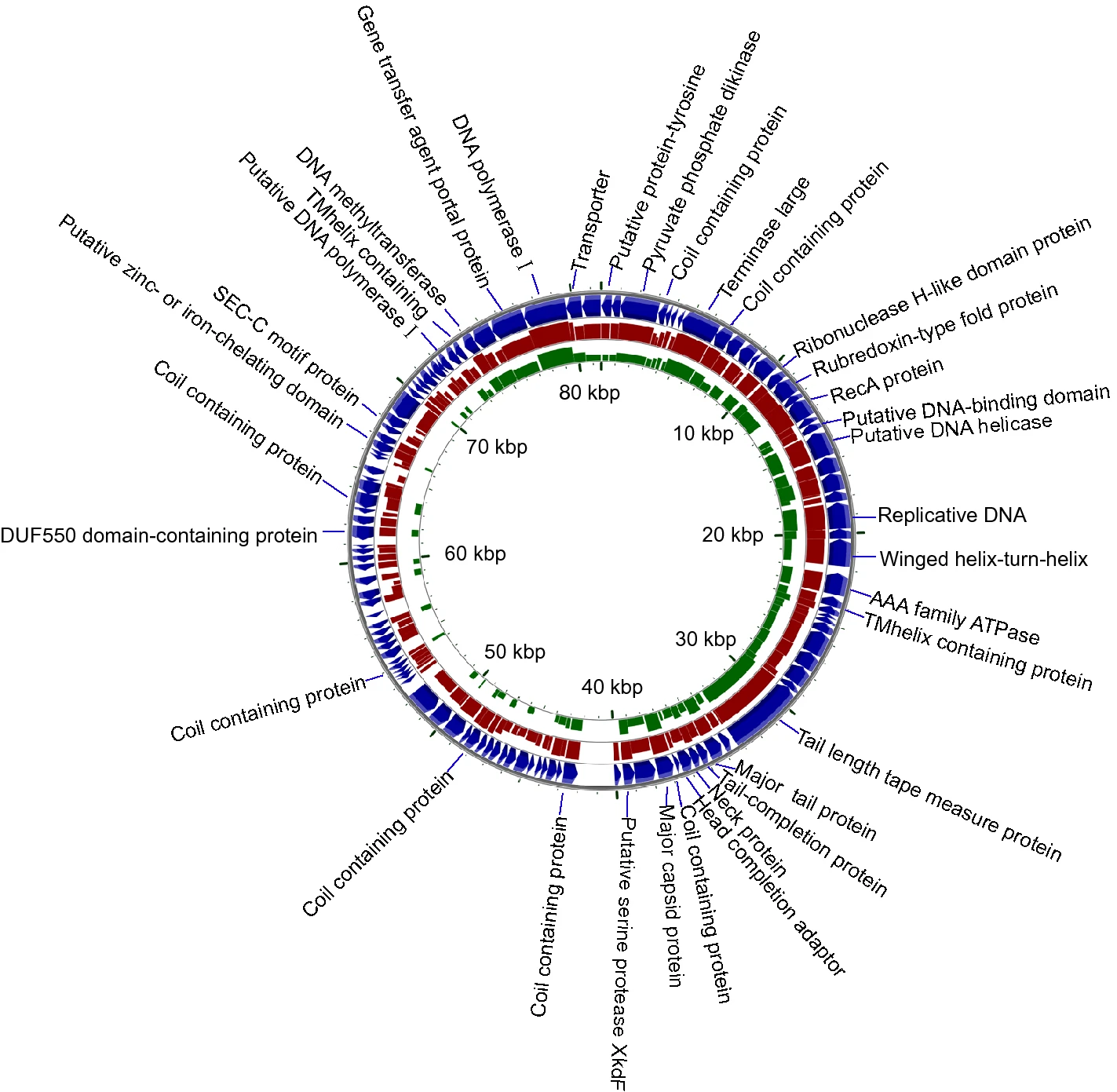
A Novel Vibriophage vB_VcaS_HC Containing Lysogeny-Related Gene Has Strong Lytic Ability against Pathogenic Bacteria
2021, 36(2): 281 doi: 10.1007/s12250-020-00271-w
To avoid the negative effects of antibiotics, using phage to prevent animal disease becomes a promising method in aquaculture. Here, a lytic phage provisionally named vB_VcaS_HC that can infect the pathogen (i.e., Vibrio campbellii 18) of prawn was isolated. The phage has an isometric head and a non-contractile tail. During phage infection, the induced host mortality in 5.5 h reached ca. 96%, with a latent period of 1.5 h and a burst size of 172 PFU/cell. It has an 81, 566 bp circular dsDNA genome containing 121 open reading frames (ORFs), and ca. 71% of the ORFs are functionally unknown. Comparative genomic and phylogenetic analysis revealed that it is a novel phage belonging to Delepquintavirus, Siphoviridae, Caudovirales. In the phage genome, besides the ordinary genes related to structure assembly and DNA metabolism, there are 10 auxiliary metabolic genes. For the first time, the pyruvate phosphate dikinase (PPDK) gene was found in phages whose product is a key rate-limiting enzyme involving Embden-Meyerhof-Parnas (EMP) reaction. Interestingly, although the phage has a strong bactericidal activity and contains a potential lysogeny related gene, i.e., the recombinase (RecA) gene, we did not find the phage turned into a lysogenic state. Meanwhile, the phage genome does not contain any bacterial virulence gene or antimicrobial resistance gene. This study represents the first comprehensive characterization of a lytic V. campbellii phage and indicates that it is a promising candidate for the treatment of V. campbellii infections.
Genetic and Molecular Characterization of H9N2 Avian Influenza Viruses Isolated from Live Poultry Markets in Hubei Province, Central China, 2013–2017
2021, 36(2): 291 doi: 10.1007/s12250-020-00260-z
H9N2 subtype avian influenza virus (AIV) is an influenza A virus that is widely spread throughout Asia, where it jeopardizes the poultry industry and provides genetic material for emerging human pathogens. To better understand the epidemicity and genetics of H9 subtype AIVs, we conducted active surveillance in live poultry markets (LPMs) in Hubei Province from 2013 to 2017. A total of 4798 samples were collected from apparent healthy poultry and environment. Real-time RT-PCR revealed that the positivity rate of influenza A was 26.6% (1275/4798), of which the H9 subtype accounted for 50.3% (641/1275) of the positive samples. Of the 132 H9N2 viral strains isolated, 48 representative strains were subjected to evolutionary analysis and genotyping. Phylogenetic analysis revealed that all H9N2 viral genes had 91.1%–100% nucleotide homology, clustered with genotype 57, and had high homology with human H9N2 viruses isolated from 2013 to 2017 in China. Using a nucleotide divergence cutoff of 95%, we identified ten distinct H9N2 genotypes that continued to change over time. Molecular analysis demonstrated that six H9N2 isolates had additional potential glycosylation sites at position 218 in the hemagglutinin protein, and all isolates had I155T and Q226L mutations. Moreover, 44 strains had A558V mutations in the PB2 protein and four had E627V mutations, along with H9N2 human infection strains A/Beijing/1/2016 and A/Beijing/1/2017. These results emphasize that the H9N2 influenza virus in Hubei continues to mutate and undergo mammalian adaptation changes, indicating the necessity of strengthening the surveillance of the AIV H9N2 subtype in LPMs.
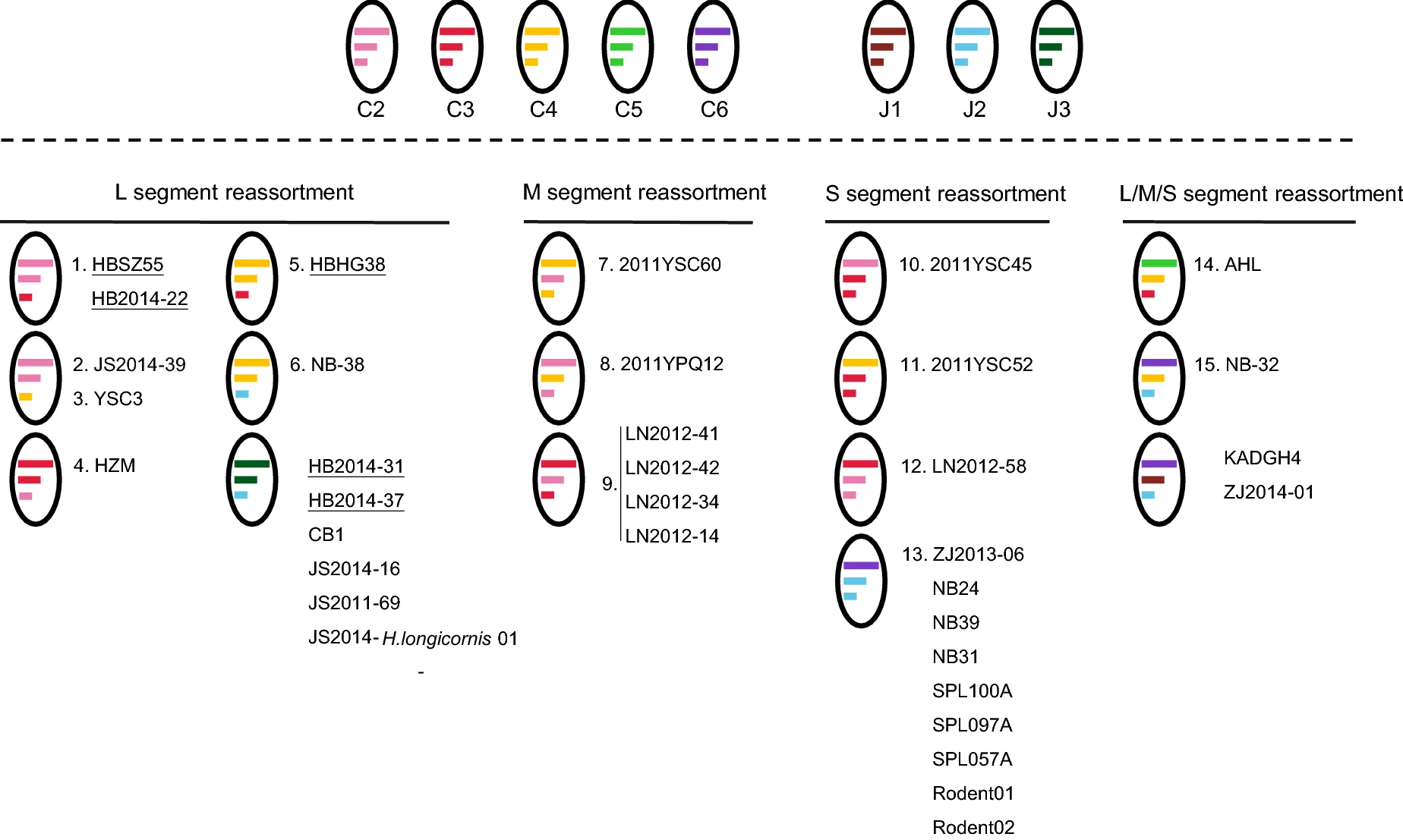
Novel SFTSV Phylogeny Reveals New Reassortment Events and Migration Routes
2021, 36(2): 300 doi: 10.1007/s12250-020-00289-0
Severe fever with thrombocytopenia syndrome virus (SFTSV), the causative agent of a febrile human disease, was first identified from central and eastern provinces in China, and later in Japan and South Korea. Hubei Province is one of the major SFTS epidemic areas in the central part of China. This study reported the isolation of 11 new SFTSV strains from patients in Hubei Province collected in 2017. Extensive phylogenetic analyses were conducted based on the complete coding sequences of SFTSV segments including the new strains. It was suggested that five different SFTSV genotypes were circulating in Hubei, and 15 reassortment patterns and migration pathways correlated with each genotype were identified, which was more than previously recognized. Hubei Province was more involved in the evolutionary events of SFTSV than that previously thought in which the evolutionary events of SFTSV were reported to be independent from those in other epidemic regions. Further divergence of SFTSV strains was suggested by pairwise comparison of SFTSV sequences from each genotype and sequence identity normalized to representative strain in genotype C1. Subsequently, amino acid variations specific for genotype(s), strain(s), or cluster(s) were inspected, which may be related to differential biological activity of SFTSV strains/genotypes. In conclusion, we analyzed the current status of SFTSV phylogeny in Hubei Province and discussed the possible events correlated to SFTSV evolution. It provided an in-depth insight into SFTSV evolution, raising concerns for the use of proper SFTSV strains in future studies.
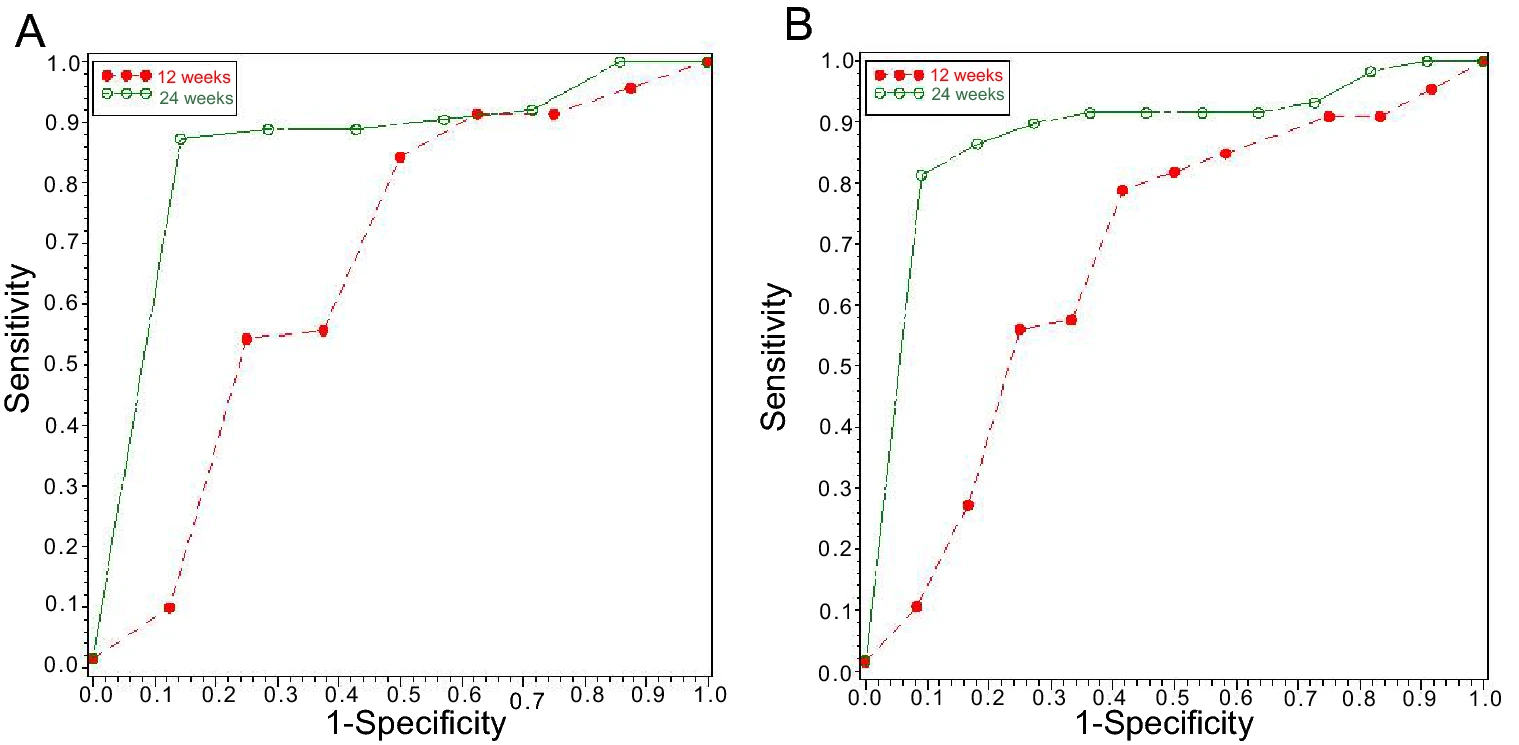
Early Serum HBsAg Kinetics as Predictor of HBsAg Loss in Patients with HBeAg-Negative Chronic Hepatitis B after Treatment with Pegylated Interferonα-2a
2021, 36(2): 311 doi: 10.1007/s12250-020-00290-7
Hepatitis B surface antigen (HBsAg) loss is an ideal treatment endpoint for patients with chronic hepatitis B (CHB). We investigated the predictive value of on-treatment HBsAg levels for HBsAg loss in hepatitis B e antigen (HBeAg)-negative CHB patients who received 120-week PEG-IFNα-2a treatment. Serum HBV DNA, HBsAg, and anti-HBs levels were assayed at baseline and every 3 months during the treatment. Of 81 patients, 12 achieved HBsAg loss, 20 achieved HBsAg < 100 IU/mL, and 49 maintained HBsAg≥100 IU/mL. HBsAg loss rate was only 3.7% at 48 weeks, while it reached to 11.1% and 14.8% after treatment of 96 weeks and 120 weeks. The cutoff HBsAg levels at 12 weeks predicting HBsAg loss at 96 weeks and 120 weeks of treatment were 400 IU/mL and 750 IU/mL, with AUC 0.725 and 0.722, positive predictive value (PPV) 29.41% and 30.56%, and negative predictive value (NPV) 93.75% and 97.78%, respectively. The cutoff HBsAg levels at 24 weeks predicting HBsAg loss at 96 weeks and 120 weeks of treatment were 174 IU/mL and 236 IU/mL respectively, with AUC 0.925 and 0.922, PPV 40.0% and 46.15%, and both NPV 100%. The predictive ability of the cutoff HBsAg levels at 24 weeks was better than that at 12 weeks for HBsAg loss at either 96 or 120 weeks (χ2=3.880, P=0.049 and χ2=4.412, P=0.036). These results indicate that extended therapy is critical to HBsAg loss in HBeAg-negative CHB patients during PEG-IFN treatment, and the HBsAg level at 24 weeks can be used to predict HBsAg loss during tailoring PEG-IFN therapy.
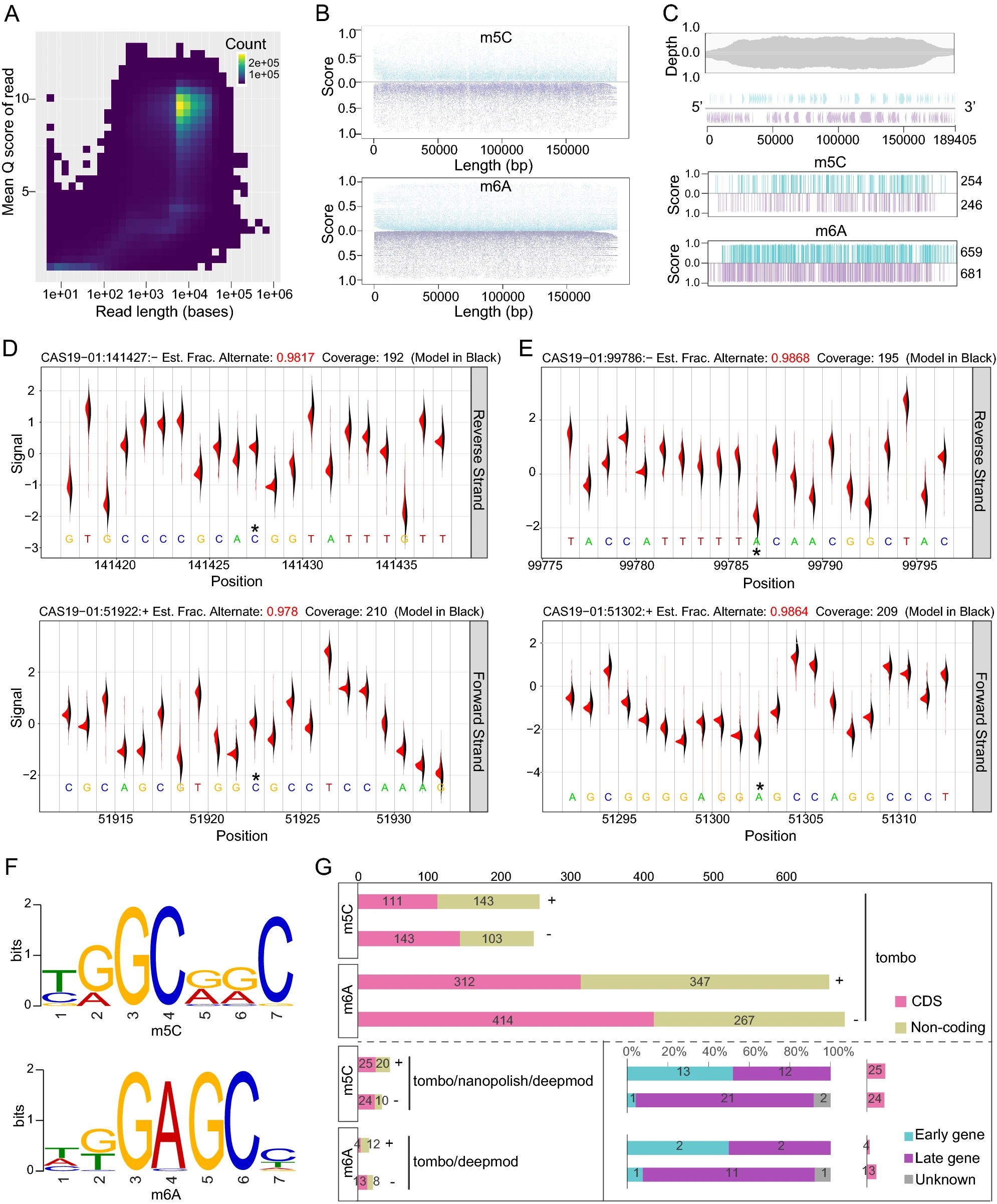
Potential m6A and m5C Methylations within the Genome of A Chinese African Swine Fever Virus Strain
2021, 36(2): 321 doi: 10.1007/s12250-020-00217-2
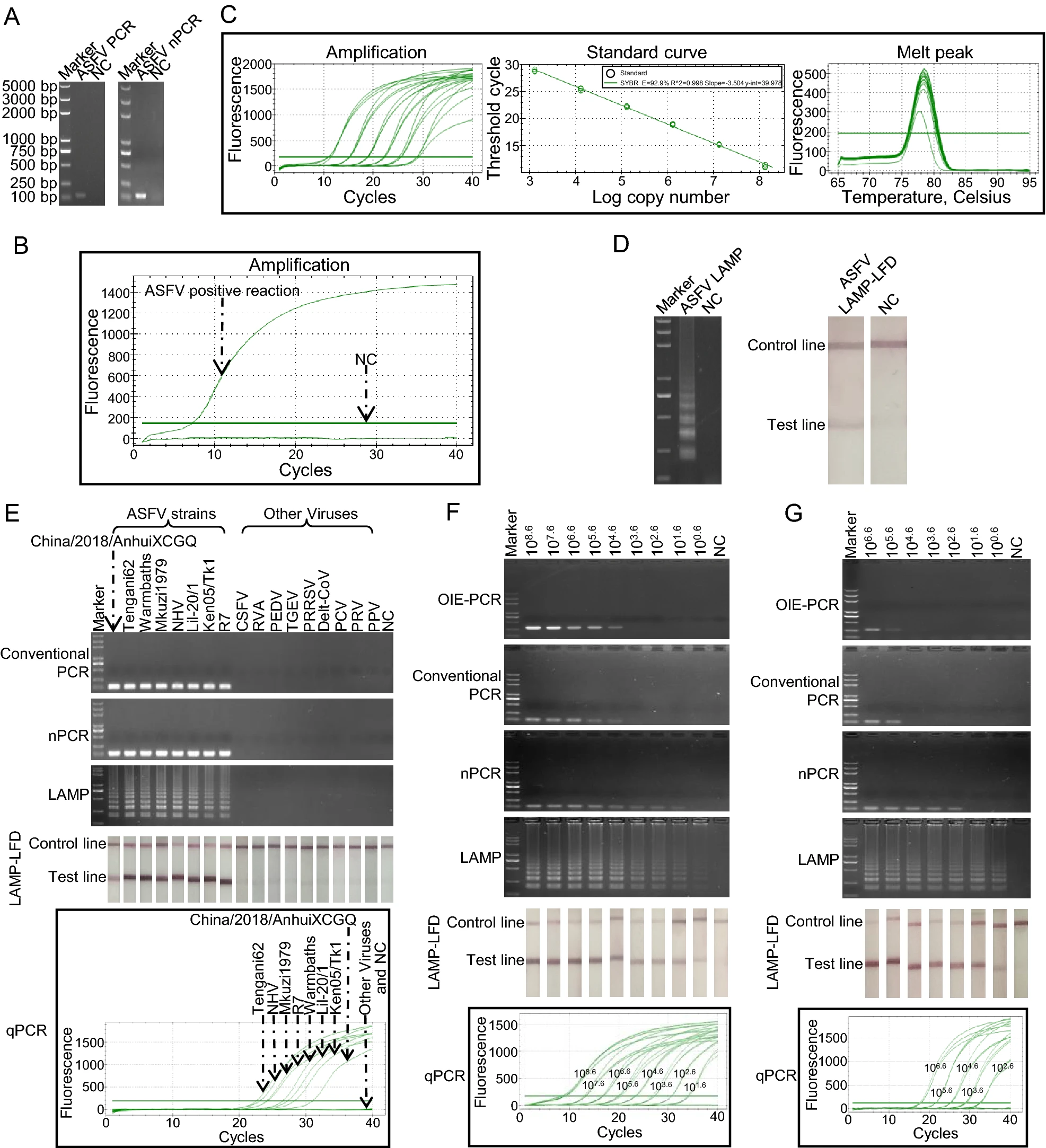
Loop-Mediated Isothermal Amplification Combined with Lateral Flow Dipstick for On-Site Diagnosis of African Swine Fever Virus
2021, 36(2): 325 doi: 10.1007/s12250-020-00309-z

First Fatal Infection and Phylodynamic Analysis of Severe Fever with Thrombocytopenia Syndrome Virus in Jilin Province, Northeastern China
2021, 36(2): 329 doi: 10.1007/s12250-020-00228-z
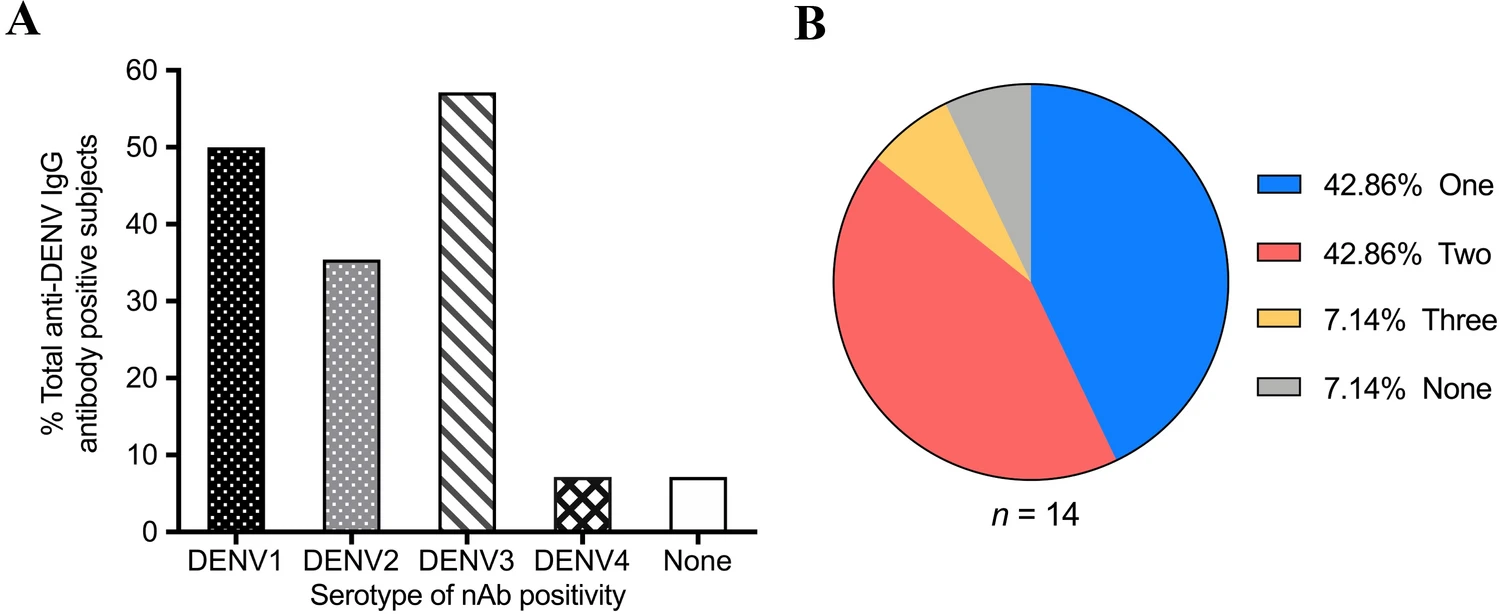
Seroprevalence of Dengue Virus among Young Adults in Beijing, China, 2019
2021, 36(2): 333 doi: 10.1007/s12250-020-00285-4

A Highly Attenuated Mumps Virus Strain of Genotype F Generated by Passaging in Vero Cells
2021, 36(2): 337 doi: 10.1007/s12250-020-00292-5

Correction to: Human Cytomegalovirus Influences Host circRNA Transcriptions during Productive Infection
2021, 36(2): 341 doi: 10.1007/s12250-020-00315-1

Correction to: A Comprehensive Review on Human Aichi Virus
2021, 36(2): 342 doi: 10.1007/s12250-020-00324-0

Correction to: Characterization of the First Genome of Porcine mastadenovirus B (HNU1 Strain) and Implications on Its Lymphoid and Special Origin
2021, 36(2): 343 doi: 10.1007/s12250-020-00328-w

















